You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001489_03302
You are here: Home > Sequence: MGYG000001489_03302
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides goldsteinii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides goldsteinii | |||||||||||
| CAZyme ID | MGYG000001489_03302 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 768975; End: 772145 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 23 | 720 | 4.4e-70 | 0.6954787234042553 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 4.67e-29 | 23 | 480 | 10 | 454 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 1.34e-20 | 24 | 464 | 11 | 443 | beta-D-glucuronidase; Provisional |
| pfam02837 | Glyco_hydro_2_N | 9.95e-16 | 25 | 156 | 1 | 135 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| PRK10340 | ebgA | 5.10e-12 | 26 | 465 | 42 | 471 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| PRK09525 | lacZ | 3.56e-10 | 20 | 455 | 47 | 477 | beta-galactosidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQD16168.1 | 0.0 | 22 | 1056 | 21 | 1075 |
| QQT56131.1 | 0.0 | 11 | 1056 | 3 | 1074 |
| QOD61956.1 | 0.0 | 3 | 1056 | 1 | 1066 |
| QTE22603.1 | 0.0 | 11 | 1056 | 9 | 1066 |
| QQT78362.1 | 0.0 | 24 | 1056 | 50 | 1096 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6S6Z_A | 3.75e-22 | 17 | 473 | 30 | 471 | Structureof beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_B Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_C Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_D Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_E Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_F Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_G Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8],6S6Z_H Structure of beta-Galactosidase from Thermotoga maritima [Thermotoga maritima MSB8] |
| 6SD0_A | 3.75e-22 | 17 | 473 | 31 | 472 | Structureof beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_B Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_C Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8],6SD0_D Structure of beta-galactosidase from Thermotoga maritima. [Thermotoga maritima MSB8] |
| 7KGY_A | 7.61e-16 | 25 | 441 | 17 | 419 | ChainA, Beta-glucuronidase [Faecalibacterium prausnitzii],7KGY_B Chain B, Beta-glucuronidase [Faecalibacterium prausnitzii],7KGY_C Chain C, Beta-glucuronidase [Faecalibacterium prausnitzii],7KGY_D Chain D, Beta-glucuronidase [Faecalibacterium prausnitzii] |
| 6ECA_A | 7.92e-16 | 26 | 441 | 41 | 439 | Lactobacillusrhamnosus Beta-glucuronidase [Lacticaseibacillus rhamnosus],6ECA_B Lactobacillus rhamnosus Beta-glucuronidase [Lacticaseibacillus rhamnosus] |
| 6JZ1_A | 9.92e-16 | 25 | 330 | 17 | 308 | Apostructure of b-glucuronidase from Ruminococcus gnavus at 1.7 Angstrom resolution [[Ruminococcus] gnavus],6JZ1_B Apo structure of b-glucuronidase from Ruminococcus gnavus at 1.7 Angstrom resolution [[Ruminococcus] gnavus],6JZ5_A b-glucuronidase from Ruminococcus gnavus in complex with D-glucuronic acid [[Ruminococcus] gnavus],6JZ5_B b-glucuronidase from Ruminococcus gnavus in complex with D-glucuronic acid [[Ruminococcus] gnavus],6JZ6_A b-glucuronidase from Ruminococcus gnavus in complex with C6-substituted uronic isofagomine [[Ruminococcus] gnavus],6JZ6_B b-glucuronidase from Ruminococcus gnavus in complex with C6-substituted uronic isofagomine [[Ruminococcus] gnavus],6JZ7_A b-glucuronidase from Ruminococcus gnavus in complex with N1-substituted uronic isofagomine [[Ruminococcus] gnavus],6JZ7_B b-glucuronidase from Ruminococcus gnavus in complex with N1-substituted uronic isofagomine [[Ruminococcus] gnavus],6JZ8_A b-glucuronidase from Ruminococcus gnavus in complex with D-glucaro 1,5-lactone [[Ruminococcus] gnavus],6JZ8_B b-glucuronidase from Ruminococcus gnavus in complex with D-glucaro 1,5-lactone [[Ruminococcus] gnavus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q56307 | 2.05e-21 | 17 | 473 | 31 | 472 | Beta-galactosidase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=lacZ PE=1 SV=2 |
| Q6D736 | 4.97e-15 | 22 | 455 | 57 | 487 | Beta-galactosidase OS=Pectobacterium atrosepticum (strain SCRI 1043 / ATCC BAA-672) OX=218491 GN=lacZ PE=3 SV=1 |
| Q03WL0 | 5.76e-14 | 23 | 455 | 43 | 471 | Beta-galactosidase OS=Leuconostoc mesenteroides subsp. mesenteroides (strain ATCC 8293 / DSM 20343 / BCRC 11652 / CCM 1803 / JCM 6124 / NCDO 523 / NBRC 100496 / NCIMB 8023 / NCTC 12954 / NRRL B-1118 / 37Y) OX=203120 GN=lacZ PE=3 SV=1 |
| A7MN76 | 7.59e-14 | 25 | 455 | 62 | 486 | Beta-galactosidase OS=Cronobacter sakazakii (strain ATCC BAA-894) OX=290339 GN=lacZ PE=3 SV=1 |
| A9MQ82 | 9.88e-14 | 21 | 455 | 48 | 477 | Beta-galactosidase OS=Salmonella arizonae (strain ATCC BAA-731 / CDC346-86 / RSK2980) OX=41514 GN=lacZ PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
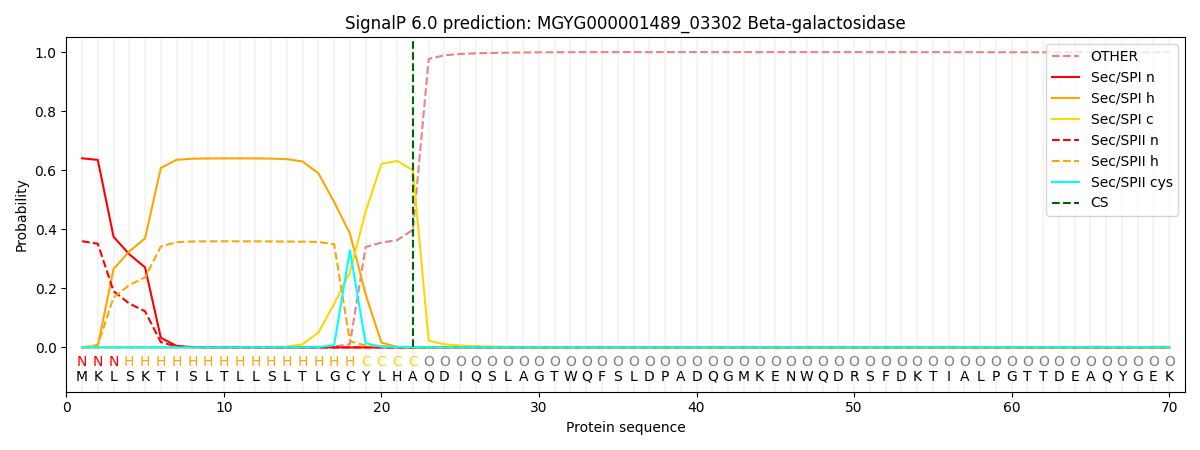
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000447 | 0.633360 | 0.365586 | 0.000197 | 0.000207 | 0.000187 |
