You are browsing environment: HUMAN GUT
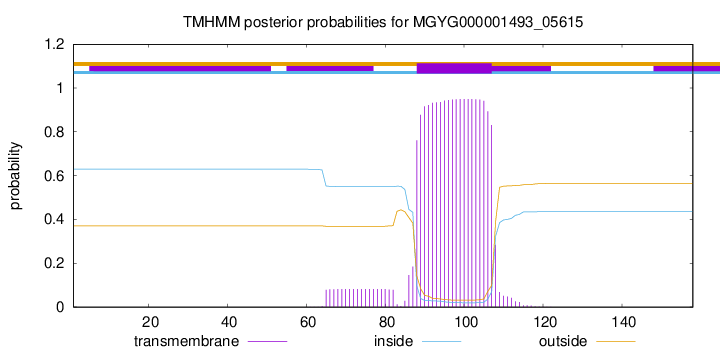
MGYG000001493_05615
Basic Information
help
Species
Enterocloster bolteae
Lineage
Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Enterocloster; Enterocloster bolteae
CAZyme ID
MGYG000001493_05615
CAZy Family
GT1
CAZyme Description
hypothetical protein
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000001493
6583702
Isolate
not provided
not provided
Gene Location
Start: 239754;
End: 240230
Strand: -
No EC number prediction in MGYG000001493_05615.
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam08660
Alg14
1.46e-14
67
157
75
171
Oligosaccharide biosynthesis protein Alg14 like. Alg14 is involved dolichol-linked oligosaccharide biosynthesis and anchors the catalytic subunit Alg13 to the ER membrane.
more
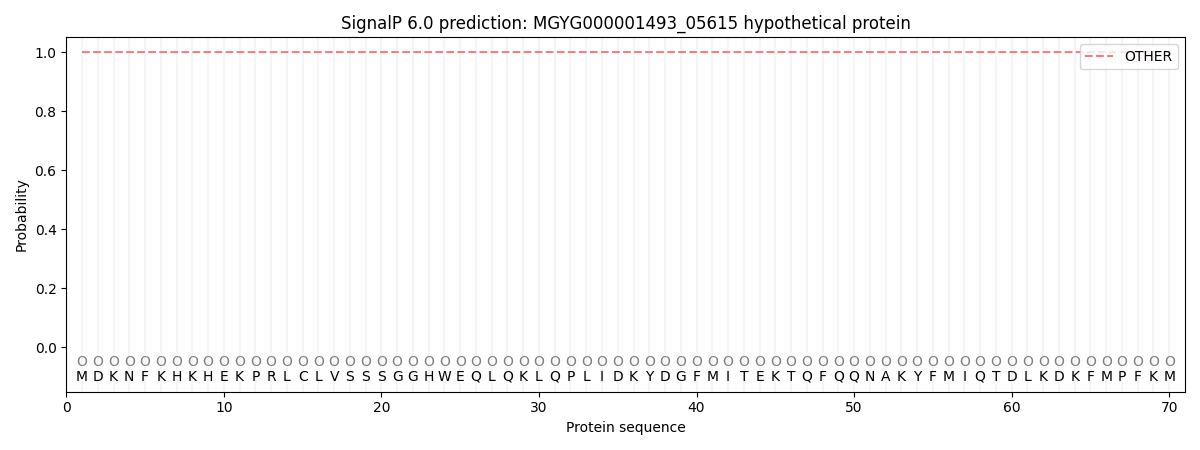
This protein is predicted as OTHER
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
1.000052
0.000000
0.000000
0.000000
0.000000
0.000000