You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001498_03836
You are here: Home > Sequence: MGYG000001498_03836
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacillus_BN testis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales_B; DSM-1321; Bacillus_BN; Bacillus_BN testis | |||||||||||
| CAZyme ID | MGYG000001498_03836 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3794790; End: 3796772 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH73 | 459 | 571 | 7.1e-20 | 0.9453125 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 3.37e-11 | 449 | 579 | 93 | 236 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam00395 | SLH | 1.91e-10 | 82 | 123 | 1 | 42 | S-layer homology domain. |
| NF033190 | inl_like_NEAT_1 | 3.93e-10 | 34 | 201 | 592 | 754 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| pfam01832 | Glucosaminidase | 1.11e-09 | 458 | 518 | 3 | 74 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| NF033190 | inl_like_NEAT_1 | 2.85e-07 | 82 | 206 | 582 | 698 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGX06524.1 | 5.48e-199 | 1 | 660 | 1 | 656 |
| QVY61327.1 | 8.07e-169 | 1 | 650 | 1 | 660 |
| QNU35619.1 | 5.50e-164 | 1 | 660 | 1 | 627 |
| QNU38948.1 | 7.00e-162 | 1 | 660 | 1 | 627 |
| QXJ38750.1 | 7.89e-161 | 1 | 660 | 1 | 627 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4Q2W_A | 7.46e-23 | 306 | 579 | 55 | 290 | CrystalStructure of pneumococcal peptidoglycan hydrolase LytB [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P59205 | 4.26e-22 | 306 | 579 | 423 | 658 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=lytB PE=1 SV=1 |
| P59206 | 4.72e-22 | 306 | 579 | 467 | 702 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=lytB PE=1 SV=1 |
| P38538 | 2.17e-12 | 1 | 133 | 31 | 173 | Surface layer protein OS=Brevibacillus choshinensis OX=54911 PE=1 SV=1 |
| P38536 | 2.49e-12 | 37 | 200 | 1694 | 1856 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| P38535 | 6.45e-12 | 37 | 203 | 920 | 1085 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
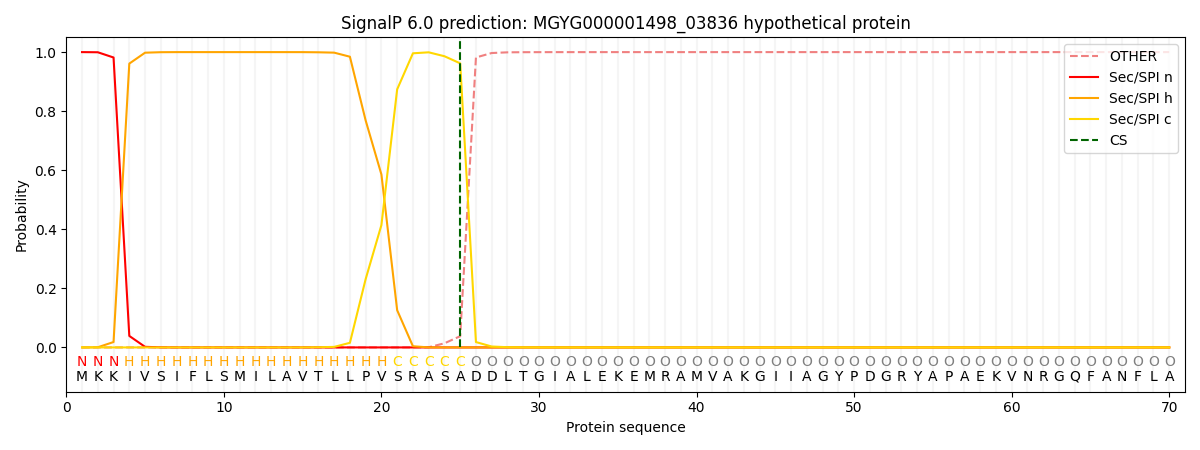
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000296 | 0.998960 | 0.000183 | 0.000186 | 0.000176 | 0.000160 |
