You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001503_00456
You are here: Home > Sequence: MGYG000001503_00456
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fermentimonas caenicola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Dysgonomonadaceae; Fermentimonas; Fermentimonas caenicola | |||||||||||
| CAZyme ID | MGYG000001503_00456 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 556571; End: 558091 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 56 | 451 | 1.1e-26 | 0.7486033519553073 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03663 | Glyco_hydro_76 | 4.24e-09 | 218 | 393 | 126 | 258 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CEA16166.1 | 0.0 | 1 | 500 | 1 | 500 |
| AWW30266.1 | 9.53e-75 | 24 | 497 | 42 | 475 |
| QDH77939.1 | 3.70e-74 | 24 | 497 | 42 | 475 |
| AGA76828.1 | 5.20e-74 | 24 | 497 | 42 | 475 |
| SCD21417.1 | 3.12e-72 | 24 | 497 | 43 | 476 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4C1S_A | 2.32e-08 | 160 | 448 | 101 | 329 | Glycosidehydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],4C1S_B Glycoside hydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
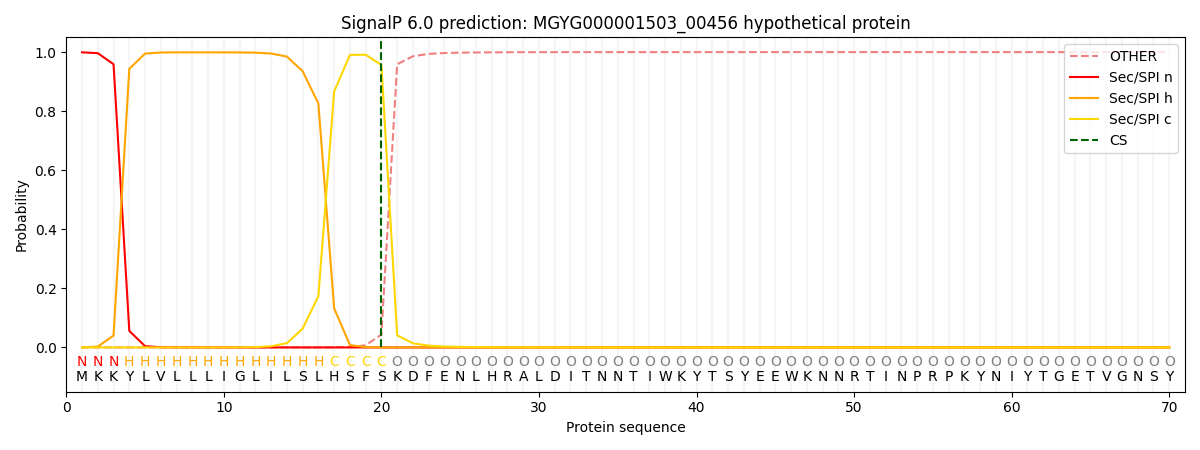
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001112 | 0.996335 | 0.001826 | 0.000220 | 0.000230 | 0.000246 |
