You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001503_01309
You are here: Home > Sequence: MGYG000001503_01309
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fermentimonas caenicola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Dysgonomonadaceae; Fermentimonas; Fermentimonas caenicola | |||||||||||
| CAZyme ID | MGYG000001503_01309 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 387214; End: 388386 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 31 | 362 | 1.1e-62 | 0.9050279329608939 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03663 | Glyco_hydro_76 | 2.04e-27 | 121 | 325 | 72 | 293 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| COG1331 | YyaL | 5.27e-09 | 125 | 282 | 401 | 561 | Uncharacterized conserved protein YyaL, SSP411 family, contains thoiredoxin and six-hairpin glycosidase-like domains [General function prediction only]. |
| COG4833 | COG4833 | 5.59e-09 | 134 | 300 | 90 | 253 | Predicted alpha-1,6-mannanase, GH76 family [Carbohydrate transport and metabolism]. |
| pfam07944 | Glyco_hydro_127 | 2.38e-06 | 148 | 332 | 69 | 250 | Beta-L-arabinofuranosidase, GH127. One member of this family, from Bidobacterium longicum, UniProtKB:E8MGH8, has been characterized as an unusual beta-L-arabinofuranosidase enzyme, EC:3.2.1.185. It rleases l-arabinose from the l-arabinofuranose (Araf)-beta1,2-Araf disaccharide and also transglycosylates 1-alkanols with retention of the anomeric configuration. Terminal beta-l-arabinofuranosyl residues have been found in arabinogalactan proteins from a mumber of different plantt species. beta-l-Arabinofuranosyl linkages with 1-4 arabinofuranosides are also found in the sugar chains of extensin and solanaceous lectins, hydroxyproline (Hyp)2-rich glycoproteins that are widely observed in plant cell wall fractions. The critical residue for catalytic activity is Glu-338, in a ET/SCAS sequence context. |
| cd04791 | LanC_SerThrkinase | 1.47e-05 | 140 | 352 | 92 | 290 | Lanthionine synthetase C-like domain associated with serine/threonine kinases. Some members of this subgroup lack the zinc binding site and the active site residues, and therefore are most likely inactive. The function of this domain is unknown. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT25297.1 | 6.35e-228 | 24 | 388 | 28 | 392 |
| QDM09503.1 | 6.58e-228 | 24 | 388 | 28 | 392 |
| CBK66336.1 | 6.58e-228 | 24 | 388 | 28 | 392 |
| QUT30759.1 | 7.38e-227 | 24 | 388 | 28 | 392 |
| QDH57402.1 | 1.09e-226 | 24 | 388 | 28 | 392 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4MU9_A | 1.42e-67 | 25 | 377 | 3 | 358 | Crystalstructure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482],4MU9_B Crystal structure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
| 4C1S_A | 2.88e-37 | 77 | 387 | 45 | 373 | Glycosidehydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],4C1S_B Glycoside hydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
| 6U4Z_A | 4.26e-32 | 77 | 387 | 169 | 498 | CrystalStructure of a family 76 glycoside hydrolase from a bovine Bacteroides thetaiotaomicron strain [Bacteroides thetaiotaomicron] |
| 4V1S_A | 1.15e-28 | 62 | 382 | 68 | 388 | Structureof the GH76 alpha-mannanase BT2949 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],4V1S_B Structure of the GH76 alpha-mannanase BT2949 from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
| 4V1R_A | 2.51e-26 | 62 | 377 | 68 | 382 | Structureof a selenomethionine derivative of the GH76 alpha- mannanase BT2949 Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482],4V1R_B Structure of a selenomethionine derivative of the GH76 alpha- mannanase BT2949 Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
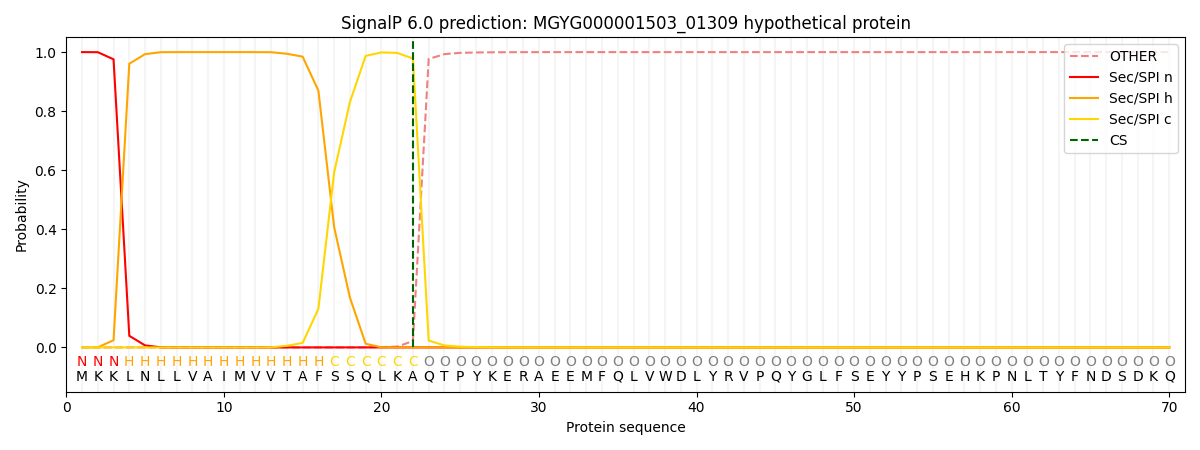
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000206 | 0.999137 | 0.000199 | 0.000154 | 0.000142 | 0.000128 |
