You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001503_01396
You are here: Home > Sequence: MGYG000001503_01396
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fermentimonas caenicola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Dysgonomonadaceae; Fermentimonas; Fermentimonas caenicola | |||||||||||
| CAZyme ID | MGYG000001503_01396 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 498222; End: 499880 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16586 | DUF5060 | 2.77e-07 | 31 | 105 | 1 | 68 | Domain of unknown function (DUF5060). This is the N-terminal domain of a putative glycoside hydrolase, DUF4038. It is found in a number of different bacterial orders. |
| COG2730 | BglC | 2.60e-04 | 142 | 419 | 39 | 319 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| COG3934 | COG3934 | 5.49e-04 | 191 | 330 | 39 | 175 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQT46981.1 | 3.58e-220 | 37 | 549 | 36 | 555 |
| QQT60502.1 | 4.05e-220 | 39 | 549 | 2 | 519 |
| BBL01046.1 | 4.73e-220 | 9 | 550 | 4 | 549 |
| QRQ60221.1 | 1.02e-219 | 37 | 549 | 36 | 555 |
| BBL06281.1 | 7.29e-214 | 35 | 550 | 33 | 548 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4LYP_A | 2.32e-07 | 175 | 399 | 76 | 350 | CrystalStructure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei [Rhizomucor miehei],4LYP_B Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei [Rhizomucor miehei] |
| 4LYQ_A | 1.24e-06 | 175 | 399 | 76 | 350 | CrystalStructure of Glycoside Hydrolase Family 5 Mannosidase from Rhizomucor miehei, E202A mutant [Rhizomucor miehei],4NRR_A Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannosyl-fructose [Rhizomucor miehei],4NRR_B Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannosyl-fructose [Rhizomucor miehei],4NRS_A Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannobiose [Rhizomucor miehei],4NRS_B Crystal Structure of Glycoside Hydrolase Family 5 Mannosidase (E202A mutant) from Rhizomucor miehei in complex with mannobiose [Rhizomucor miehei] |
| 4LYR_A | 1.24e-06 | 175 | 399 | 76 | 350 | GlycosideHydrolase Family 5 Mannosidase from Rhizomucor miehei, E301A mutant [Rhizomucor miehei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q5AZ53 | 1.50e-06 | 259 | 336 | 159 | 239 | Mannan endo-1,4-beta-mannosidase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
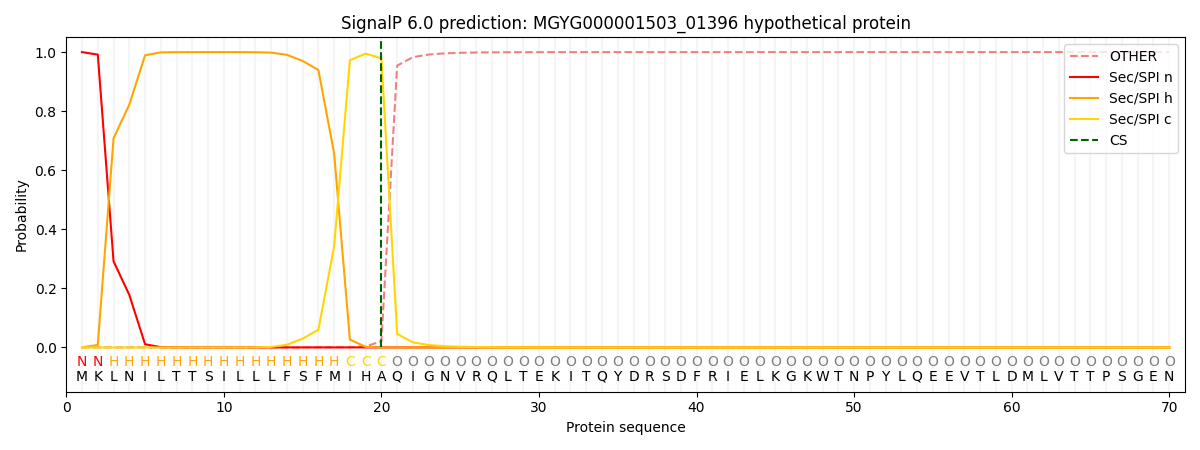
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000478 | 0.998731 | 0.000248 | 0.000177 | 0.000165 | 0.000169 |
