You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001503_01685
You are here: Home > Sequence: MGYG000001503_01685
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fermentimonas caenicola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Dysgonomonadaceae; Fermentimonas; Fermentimonas caenicola | |||||||||||
| CAZyme ID | MGYG000001503_01685 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 849273; End: 850418 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 34 | 366 | 2.3e-90 | 0.9385474860335196 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4833 | COG4833 | 7.10e-64 | 41 | 372 | 3 | 368 | Predicted alpha-1,6-mannanase, GH76 family [Carbohydrate transport and metabolism]. |
| pfam03663 | Glyco_hydro_76 | 1.46e-42 | 41 | 365 | 2 | 348 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| cd04434 | LanC_like | 1.19e-04 | 87 | 232 | 62 | 210 | Cyclases involved in the biosynthesis of lantibiotics, and similar proteins. LanC is the cyclase enzyme of the lanthionine synthetase. Lanthionine is a lantibiotic, a unique class of peptide antibiotics. They are ribosomally synthesized as a precursor peptide and then post-translationally modified to contain thioether cross-links called lanthionines (Lans) or methyllanthionines (MeLans), in addition to 2,3-didehydroalanine (Dha) and (Z)-2,3-didehydrobutyrine (Dhb). These unusual amino acids are introduced by the dehydration of serine and threonine residues, followed by thioether formation via addition of cysteine thiols, catalysed by LanB and LanC or LanM. LanC, the cyclase component, is a zinc metalloprotein, whose bound metal has been proposed to activate the thiol substrate for nucleophilic addition. A related domain is also present in LanM and other pro- and eukaryotic proteins with poorly characterized functions. |
| cd04791 | LanC_SerThrkinase | 0.001 | 129 | 299 | 92 | 248 | Lanthionine synthetase C-like domain associated with serine/threonine kinases. Some members of this subgroup lack the zinc binding site and the active site residues, and therefore are most likely inactive. The function of this domain is unknown. |
| cd04792 | LanM-like | 0.002 | 129 | 216 | 657 | 743 | Cyclases involved in the biosynthesis of class II lantibiotics, and similar proteins. LanM-like proteins. LanM is a bifunctional enzyme, involved in the synthesis of class II lantibiotics. It is responsible for both the dehydration and the cyclization of the precursor-peptide during lantibiotic synthesis. The C-terminal domain shows similarity to LanC, the cyclase component of the lan operon, but the N terminus seems to be unrelated to the dehydratase, LanB. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CEA14963.1 | 5.32e-289 | 1 | 381 | 1 | 381 |
| AHW60278.1 | 9.80e-179 | 3 | 381 | 27 | 400 |
| QGY43662.1 | 4.98e-164 | 1 | 380 | 1 | 377 |
| QIA07428.1 | 2.04e-161 | 1 | 381 | 1 | 380 |
| AXY74790.1 | 9.31e-134 | 18 | 378 | 17 | 382 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6SHD_A | 4.12e-104 | 41 | 375 | 54 | 390 | Structureof the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_B Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_C Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6] |
| 6Y8F_A | 1.37e-102 | 41 | 375 | 55 | 391 | ChainA, Alpha-1,6-endo-mannanase GH76A mutant [Salegentibacter sp. Hel_I_6] |
| 6SHM_A | 1.10e-101 | 41 | 375 | 55 | 391 | Aninactive (D136A and D137A) variant of alpha-1,6-mannanase, GH76A of Salegentibacter sp. HEL1_6 in complex with alpha-1,6-mannotetrose [Salegentibacter sp. Hel_I_6] |
| 3K7X_A | 5.59e-67 | 71 | 378 | 40 | 344 | ChainA, Lin0763 protein [Listeria innocua] |
| 4BOK_A | 1.22e-35 | 51 | 348 | 13 | 317 | ChainA, Alpha-1,6-mannanase [Niallia circulans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q75DG6 | 5.26e-10 | 74 | 376 | 63 | 391 | Mannan endo-1,6-alpha-mannosidase DCW1 OS=Ashbya gossypii (strain ATCC 10895 / CBS 109.51 / FGSC 9923 / NRRL Y-1056) OX=284811 GN=DCW1 PE=3 SV=2 |
| P36091 | 1.58e-07 | 63 | 329 | 46 | 340 | Mannan endo-1,6-alpha-mannosidase DCW1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=DCW1 PE=1 SV=1 |
| Q5AD78 | 3.72e-07 | 109 | 370 | 103 | 386 | Mannan endo-1,6-alpha-mannosidase DCW1 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=DCW1 PE=1 SV=1 |
| Q6FLP9 | 4.90e-07 | 62 | 303 | 48 | 314 | Mannan endo-1,6-alpha-mannosidase DCW1 OS=Candida glabrata (strain ATCC 2001 / CBS 138 / JCM 3761 / NBRC 0622 / NRRL Y-65) OX=284593 GN=DCW1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
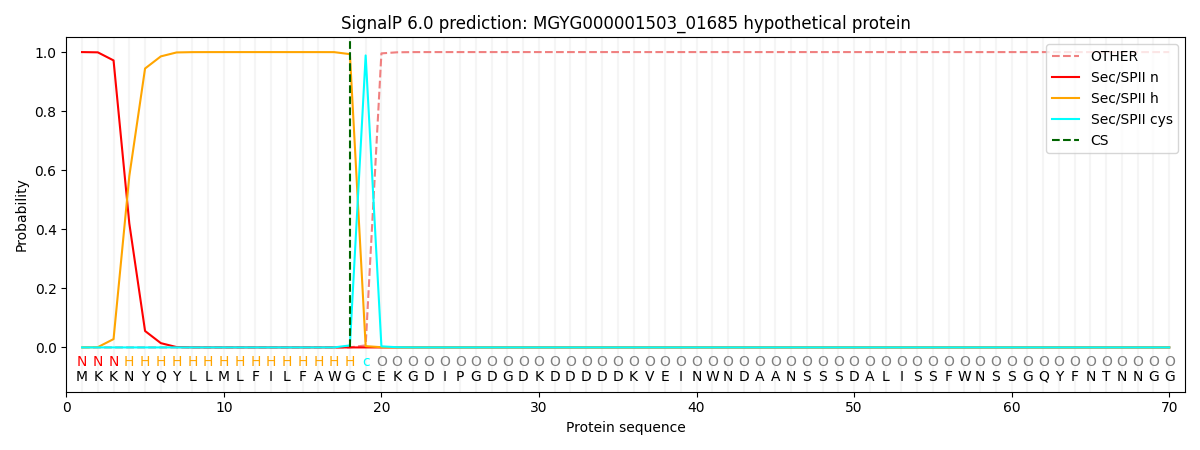
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000016 | 1.000040 | 0.000000 | 0.000000 | 0.000000 |
