You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001503_02171
You are here: Home > Sequence: MGYG000001503_02171
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fermentimonas caenicola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Dysgonomonadaceae; Fermentimonas; Fermentimonas caenicola | |||||||||||
| CAZyme ID | MGYG000001503_02171 | |||||||||||
| CAZy Family | GH55 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1412687; End: 1415719 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH55 | 47 | 630 | 4.4e-83 | 0.7864864864864864 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam12708 | Pectate_lyase_3 | 1.50e-11 | 55 | 269 | 12 | 213 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| pfam12708 | Pectate_lyase_3 | 7.08e-10 | 383 | 492 | 1 | 121 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| COG3386 | YvrE | 2.14e-04 | 744 | 1003 | 37 | 277 | Sugar lactone lactonase YvrE [Carbohydrate transport and metabolism]. |
| COG5434 | Pgu1 | 5.88e-04 | 384 | 511 | 83 | 208 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| cd05819 | NHL | 0.009 | 722 | 789 | 138 | 211 | NHL repeat unit of beta-propeller proteins. The NHL(NCL-1, HT2A and LIN-41)-repeat is found in multiple tandem copies, typically as 6 instances. It is about 40 residues long and resembles the WD repeat and other beta-propeller structures. The repeats have a catalytic activity in Peptidyl-glycine alpha-amidating monooxygenase; proteolysis has shown that the Peptidyl-alpha-hydroxyglycine alpha-amidating lyase (PAL) activity is localized to the repeats. Tripartite motif-containing protein 32 interacts with the activation domain of Tat. This interaction is mediated by the NHL repeats. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIA08652.1 | 0.0 | 26 | 1009 | 17 | 984 |
| QUT68913.1 | 0.0 | 31 | 1010 | 30 | 1000 |
| QUT62546.1 | 0.0 | 31 | 1010 | 30 | 1000 |
| BBK89257.1 | 0.0 | 31 | 1010 | 24 | 994 |
| QQA31096.1 | 0.0 | 31 | 1010 | 30 | 1000 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3EQN_A | 3.19e-14 | 48 | 613 | 53 | 630 | ChainA, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium],3EQN_B Chain B, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium],3EQO_A Chain A, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium],3EQO_B Chain B, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium] |
| 5M5Z_A | 5.38e-09 | 79 | 470 | 71 | 470 | ChainA, Beta-1,3-glucanase [Thermochaetoides thermophila],5M60_A Chain A, Beta-1,3-glucanase [Thermochaetoides thermophila] |
| 7CHU_A | 1.37e-06 | 369 | 453 | 62 | 158 | ChainA, Putative pectin lyase [Geobacillus virus E2],7CHU_B Chain B, Putative pectin lyase [Geobacillus virus E2],7CHU_C Chain C, Putative pectin lyase [Geobacillus virus E2] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P49426 | 1.96e-09 | 22 | 671 | 45 | 703 | Glucan 1,3-beta-glucosidase OS=Cochliobolus carbonum OX=5017 GN=EXG1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
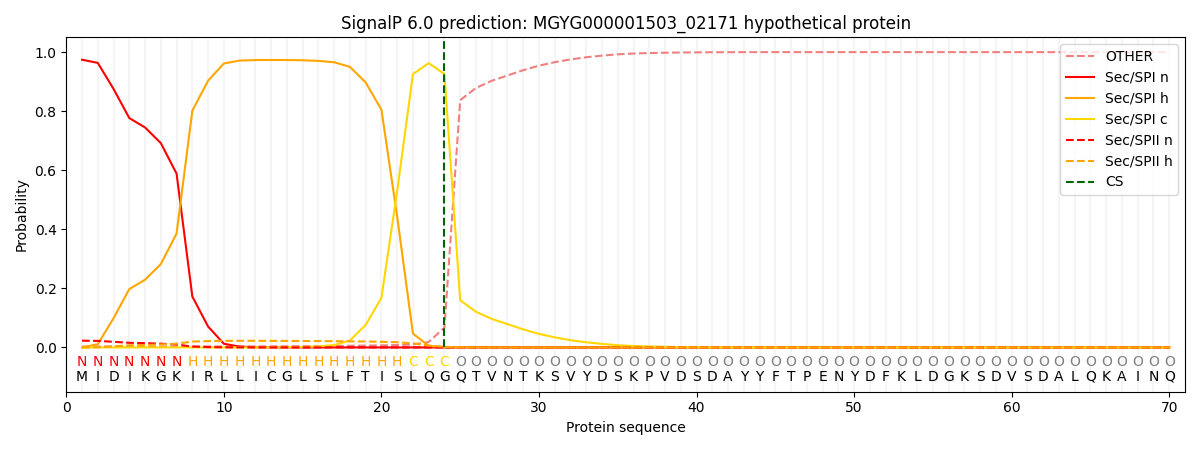
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.004900 | 0.969682 | 0.024543 | 0.000323 | 0.000271 | 0.000259 |
