You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001503_02205
You are here: Home > Sequence: MGYG000001503_02205
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fermentimonas caenicola | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Dysgonomonadaceae; Fermentimonas; Fermentimonas caenicola | |||||||||||
| CAZyme ID | MGYG000001503_02205 | |||||||||||
| CAZy Family | GH109 | |||||||||||
| CAZyme Description | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1458744; End: 1460018 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH109 | 32 | 225 | 1e-25 | 0.46115288220551376 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG0673 | MviM | 1.97e-35 | 34 | 347 | 4 | 290 | Predicted dehydrogenase [General function prediction only]. |
| pfam01408 | GFO_IDH_MocA | 7.30e-15 | 34 | 156 | 1 | 108 | Oxidoreductase family, NAD-binding Rossmann fold. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
| pfam02894 | GFO_IDH_MocA_C | 1.04e-05 | 183 | 395 | 2 | 197 | Oxidoreductase family, C-terminal alpha/beta domain. This family of enzymes utilize NADP or NAD. This family is called the GFO/IDH/MOCA family in swiss-prot. |
| PRK11579 | PRK11579 | 3.03e-04 | 105 | 243 | 59 | 183 | putative oxidoreductase; Provisional |
| PRK10206 | PRK10206 | 0.002 | 105 | 215 | 59 | 172 | putative oxidoreductase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CEA16880.1 | 5.21e-313 | 2 | 424 | 1 | 423 |
| BBD45631.1 | 8.85e-250 | 2 | 424 | 1 | 423 |
| SCD19022.1 | 1.72e-240 | 2 | 424 | 1 | 423 |
| BBD45948.1 | 2.93e-188 | 4 | 424 | 6 | 426 |
| QEC68724.1 | 3.39e-165 | 3 | 424 | 5 | 426 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4N54_A | 1.21e-16 | 99 | 398 | 66 | 346 | ChainA, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4N54_B Chain B, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4N54_C Chain C, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4N54_D Chain D, Inositol dehydrogenase [Lacticaseibacillus casei BL23] |
| 4MKX_A | 1.24e-16 | 99 | 398 | 69 | 349 | ChainA, Inositol dehydrogenase [Lacticaseibacillus casei BL23],4MKZ_A Chain A, Inositol dehydrogenase [Lacticaseibacillus casei BL23] |
| 3CEA_A | 2.80e-16 | 81 | 266 | 42 | 215 | ChainA, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1],3CEA_B Chain B, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1],3CEA_C Chain C, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1],3CEA_D Chain D, Myo-inositol 2-dehydrogenase [Lactiplantibacillus plantarum WCFS1] |
| 6O15_A | 7.07e-13 | 34 | 283 | 2 | 221 | Crystalstructure of a putative oxidoreductase YjhC from Escherichia coli in complex with NAD(H) [Escherichia coli K-12],6O15_B Crystal structure of a putative oxidoreductase YjhC from Escherichia coli in complex with NAD(H) [Escherichia coli K-12] |
| 4HKT_A | 9.73e-13 | 34 | 254 | 4 | 189 | Crystalstructure of a putative myo-inositol dehydrogenase from Sinorhizobium meliloti 1021 (Target PSI-012312) [Sinorhizobium meliloti 1021],4HKT_B Crystal structure of a putative myo-inositol dehydrogenase from Sinorhizobium meliloti 1021 (Target PSI-012312) [Sinorhizobium meliloti 1021],4HKT_C Crystal structure of a putative myo-inositol dehydrogenase from Sinorhizobium meliloti 1021 (Target PSI-012312) [Sinorhizobium meliloti 1021],4HKT_D Crystal structure of a putative myo-inositol dehydrogenase from Sinorhizobium meliloti 1021 (Target PSI-012312) [Sinorhizobium meliloti 1021] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q88S38 | 3.88e-15 | 35 | 313 | 8 | 254 | Inositol 2-dehydrogenase/D-chiro-inositol 3-dehydrogenase OS=Lactiplantibacillus plantarum (strain ATCC BAA-793 / NCIMB 8826 / WCFS1) OX=220668 GN=iolG PE=3 SV=1 |
| A4FID1 | 6.85e-14 | 34 | 315 | 3 | 250 | Inositol 2-dehydrogenase 2 OS=Saccharopolyspora erythraea (strain ATCC 11635 / DSM 40517 / JCM 4748 / NBRC 13426 / NCIMB 8594 / NRRL 2338) OX=405948 GN=iolG2 PE=3 SV=1 |
| O05389 | 2.27e-13 | 37 | 317 | 8 | 257 | Uncharacterized oxidoreductase YrbE OS=Bacillus subtilis (strain 168) OX=224308 GN=yrbE PE=3 SV=2 |
| O68965 | 1.22e-12 | 34 | 254 | 3 | 188 | Inositol 2-dehydrogenase OS=Rhizobium meliloti (strain 1021) OX=266834 GN=idhA PE=1 SV=2 |
| P40332 | 3.21e-12 | 105 | 310 | 62 | 251 | scyllo-inositol 2-dehydrogenase (NAD(+)) OS=Bacillus subtilis (strain 168) OX=224308 GN=iolX PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as TAT
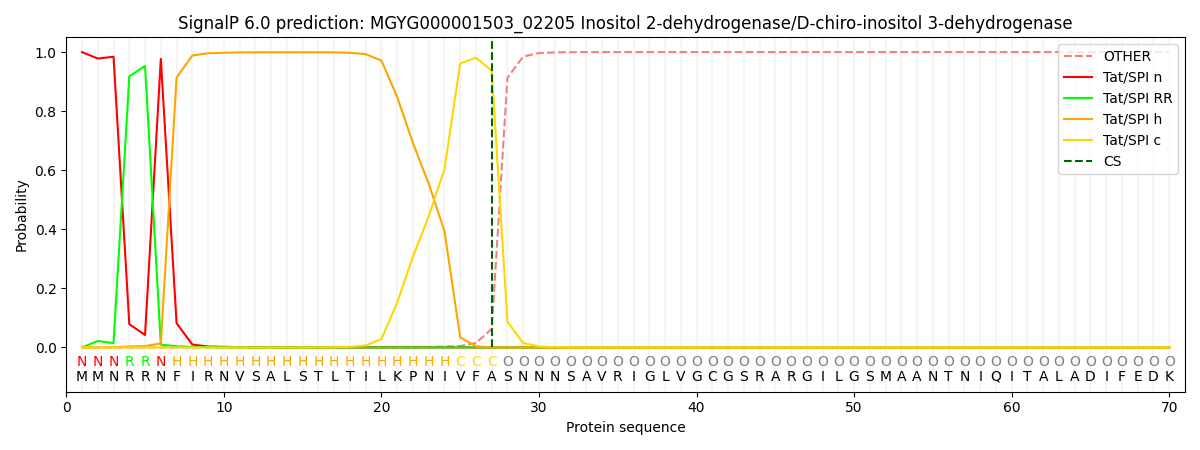
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000003 | 0.000010 | 0.000000 | 0.999974 | 0.000005 | 0.000000 |
