You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001506_00019
You are here: Home > Sequence: MGYG000001506_00019
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
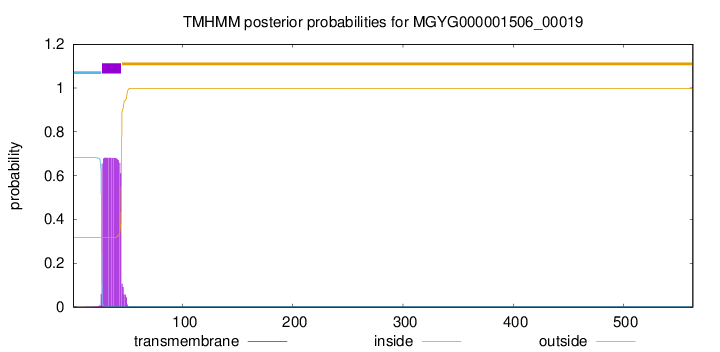
TMHMM annotations
Basic Information help
| Species | Neofamilia massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Tissierellales; Peptoniphilaceae; Neofamilia; Neofamilia massiliensis | |||||||||||
| CAZyme ID | MGYG000001506_00019 | |||||||||||
| CAZy Family | GH171 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12707; End: 14398 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH171 | 218 | 562 | 9.5e-106 | 0.9943661971830986 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07075 | DUF1343 | 2.08e-155 | 218 | 562 | 1 | 362 | Protein of unknown function (DUF1343). This family consists of several hypothetical bacterial proteins of around 400 residues in length. The function of this family is unknown. |
| COG3876 | YbbC | 3.74e-117 | 179 | 562 | 11 | 409 | Uncharacterized conserved protein YbbC, DUF1343 family [Function unknown]. |
| cd06309 | PBP1_galactofuranose_YtfQ-like | 1.18e-06 | 293 | 392 | 54 | 133 | periplasmic binding domain of ABC-type galactofuranose YtfQ-like transport systems. Periplasmic binding domain of ABC-type YtfQ-like transport systems. The YtfQ protein from Escherichia coli is up-regulated under glucose-limited conditions and shares homology with a family of pentose/hexose sugar-binding proteins of the type 1 periplasmic binding protein superfamily. Members of this group are predicted to be involved in the transport of sugar-containing molecules across cellular and organellar membranes; however their ligand specificity is not determined experimentally. |
| cd06308 | PBP1_sensor_kinase-like | 2.97e-04 | 294 | 397 | 56 | 135 | periplasmic binding domain of two-component sensor kinase signaling systems. Periplasmic binding domain of two-component sensor kinase signaling systems, some of which are fused with a C-terminal histidine kinase A domain (HisK) and/or a signal receiver domain (REC). Members of this group share homology with a family of pentose/hexose sugar-binding proteins of the type 1 periplasmic binding protein superfamily and are predicted to be involved in sensing of environmental stimuli; their substrate specificities, however, are not known in detail. |
| pfam07833 | Cu_amine_oxidN1 | 0.001 | 112 | 178 | 23 | 89 | Copper amine oxidase N-terminal domain. Copper amine oxidases catalyze the oxidative deamination of primary amines to the corresponding aldehydes, while reducing molecular oxygen to hydrogen peroxide. These enzymes are dimers of identical subunits, each comprising four domains. The N-terminal domain, which is absent in some amine oxidases, consists of a five-stranded antiparallel beta sheet twisted around an alpha helix. The D1 domains from the two subunits comprise the 'stalk' of the mushroom-shaped dimer, and interact with each other but do not pack tightly against each other. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEJ36366.1 | 9.76e-187 | 25 | 563 | 2 | 541 |
| CBH20341.1 | 4.99e-171 | 198 | 562 | 53 | 417 |
| EFE27594.2 | 1.26e-164 | 191 | 563 | 49 | 421 |
| QGU96809.1 | 1.83e-152 | 199 | 562 | 12 | 374 |
| ACB84259.1 | 8.02e-143 | 183 | 561 | 31 | 408 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4K05_A | 1.03e-68 | 192 | 563 | 8 | 399 | Crystalstructure of a DUF1343 family protein (BF0371) from Bacteroides fragilis NCTC 9343 at 1.65 A resolution [Bacteroides fragilis NCTC 9343],4K05_B Crystal structure of a DUF1343 family protein (BF0371) from Bacteroides fragilis NCTC 9343 at 1.65 A resolution [Bacteroides fragilis NCTC 9343] |
| 4JJA_A | 3.89e-35 | 197 | 563 | 6 | 369 | Crystalstructure of a DUF1343 family protein (BF0379) from Bacteroides fragilis NCTC 9343 at 1.30 A resolution [Bacteroides fragilis NCTC 9343] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P40407 | 2.34e-96 | 188 | 562 | 31 | 414 | Uncharacterized protein YbbC OS=Bacillus subtilis (strain 168) OX=224308 GN=ybbC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
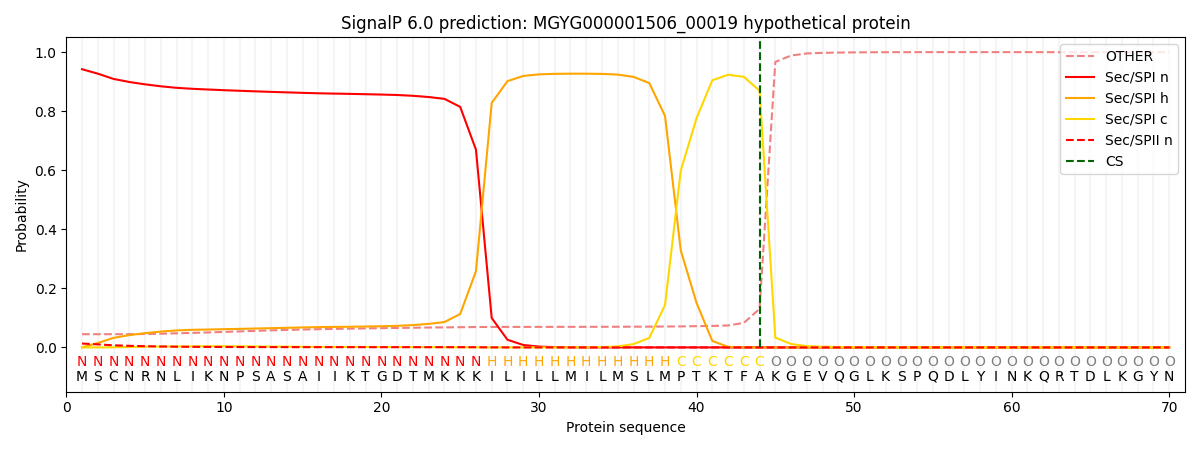
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.046924 | 0.937051 | 0.015238 | 0.000320 | 0.000229 | 0.000225 |