You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001507_02845
You are here: Home > Sequence: MGYG000001507_02845
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
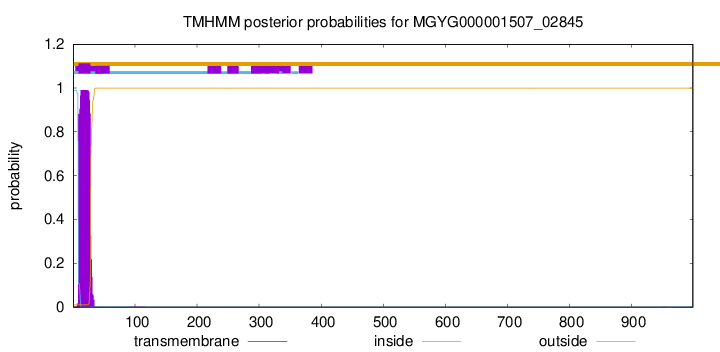
TMHMM annotations
Basic Information help
| Species | Paenibacillus ihuae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus ihuae | |||||||||||
| CAZyme ID | MGYG000001507_02845 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3009729; End: 3012728 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 515 | 815 | 1.3e-80 | 0.9966996699669967 |
| CBM23 | 831 | 990 | 1.5e-49 | 0.9753086419753086 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02156 | Glyco_hydro_26 | 2.89e-48 | 515 | 815 | 1 | 311 | Glycosyl hydrolase family 26. |
| COG4124 | ManB2 | 6.07e-20 | 640 | 794 | 160 | 323 | Beta-mannanase [Carbohydrate transport and metabolism]. |
| pfam03425 | CBM_11 | 4.01e-13 | 825 | 989 | 1 | 170 | Carbohydrate binding domain (family 11). |
| cd00063 | FN3 | 6.65e-10 | 415 | 509 | 1 | 93 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| pfam02018 | CBM_4_9 | 8.13e-10 | 36 | 157 | 2 | 134 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QSF42801.1 | 0.0 | 1 | 999 | 1 | 999 |
| CQR55720.1 | 0.0 | 1 | 997 | 1 | 998 |
| ASA19674.1 | 0.0 | 3 | 995 | 6 | 1001 |
| QQZ59662.1 | 0.0 | 7 | 680 | 35 | 709 |
| QQZ59663.1 | 7.77e-205 | 680 | 997 | 2 | 319 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q75_A | 2.77e-79 | 512 | 815 | 21 | 326 | Thestructure of GH26A from Muricauda sp. MAR_2010_75 [Muricauda sp. MAR_2010_75],6Q75_B The structure of GH26A from Muricauda sp. MAR_2010_75 [Muricauda sp. MAR_2010_75] |
| 6HPF_A | 1.57e-34 | 514 | 789 | 6 | 280 | Structureof Inactive E165Q mutant of fungal non-CBM carrying GH26 endo-b-mannanase from Yunnania penicillata in complex with alpha-62-61-di-galactosyl-mannotriose [Yunnania penicillata] |
| 3ZM8_A | 1.50e-32 | 516 | 804 | 147 | 432 | ChainA, Gh26 Endo-beta-1,4-mannanase [Podospora anserina S mat+] |
| 2WHK_A | 1.47e-29 | 514 | 824 | 5 | 333 | Structureof Bacillus subtilis mannanase man26 [Bacillus subtilis] |
| 2QHA_A | 4.09e-29 | 514 | 824 | 5 | 333 | FromStructure to Function: Insights into the Catalytic Substrate Specificity and Thermostability Displayed by Bacillus subtilis mannanase BCman [Bacillus subtilis],2QHA_B From Structure to Function: Insights into the Catalytic Substrate Specificity and Thermostability Displayed by Bacillus subtilis mannanase BCman [Bacillus subtilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| G2Q4H7 | 1.82e-34 | 516 | 806 | 176 | 466 | Mannan endo-1,4-beta-mannosidase OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=Man26A PE=1 SV=1 |
| P55296 | 1.48e-33 | 511 | 815 | 152 | 459 | Mannan endo-1,4-beta-mannosidase A OS=Piromyces sp. OX=45796 GN=MANA PE=2 SV=1 |
| P55297 | 4.46e-33 | 509 | 815 | 151 | 460 | Mannan endo-1,4-beta-mannosidase B OS=Piromyces sp. OX=45796 GN=MANB PE=2 SV=1 |
| P55298 | 1.39e-32 | 515 | 815 | 157 | 458 | Mannan endo-1,4-beta-mannosidase C OS=Piromyces sp. OX=45796 GN=MANC PE=2 SV=1 |
| Q5AWB7 | 1.27e-30 | 514 | 789 | 28 | 320 | Probable mannan endo-1,4-beta-mannosidase E OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manE PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
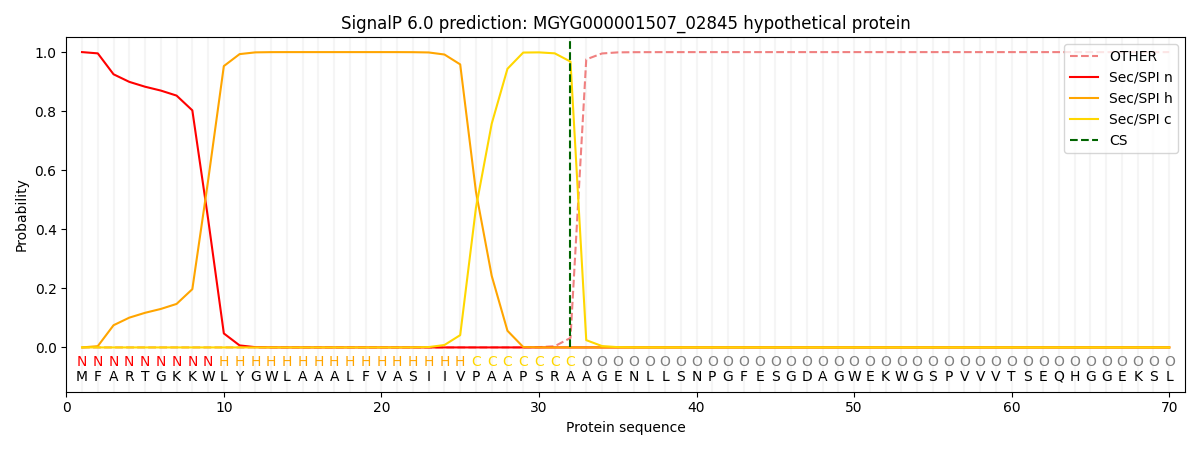
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000495 | 0.998755 | 0.000195 | 0.000215 | 0.000171 | 0.000152 |