You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001507_04469
You are here: Home > Sequence: MGYG000001507_04469
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus ihuae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus ihuae | |||||||||||
| CAZyme ID | MGYG000001507_04469 | |||||||||||
| CAZy Family | GH126 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 391077; End: 392231 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH126 | 58 | 376 | 6.9e-105 | 0.9906542056074766 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QSF47998.1 | 4.98e-247 | 13 | 383 | 1 | 371 |
| AIQ49912.1 | 1.10e-189 | 8 | 384 | 2 | 378 |
| AIQ55349.1 | 2.50e-178 | 8 | 378 | 2 | 372 |
| AIQ21110.1 | 1.59e-173 | 12 | 378 | 1 | 371 |
| AIQ32940.1 | 1.87e-171 | 1 | 378 | 1 | 382 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3REN_A | 1.06e-39 | 53 | 376 | 22 | 345 | CPF_2247,a novel alpha-amylase from Clostridium perfringens [Clostridium perfringens],3REN_B CPF_2247, a novel alpha-amylase from Clostridium perfringens [Clostridium perfringens] |
| 6R2M_A | 5.87e-12 | 91 | 318 | 49 | 266 | Crystalstructure of PssZ from Listeria monocytogenes [Listeria monocytogenes],6R2M_B Crystal structure of PssZ from Listeria monocytogenes [Listeria monocytogenes] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O31486 | 6.49e-21 | 41 | 375 | 38 | 351 | Putative lipoprotein YdaJ OS=Bacillus subtilis (strain 168) OX=224308 GN=ydaJ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
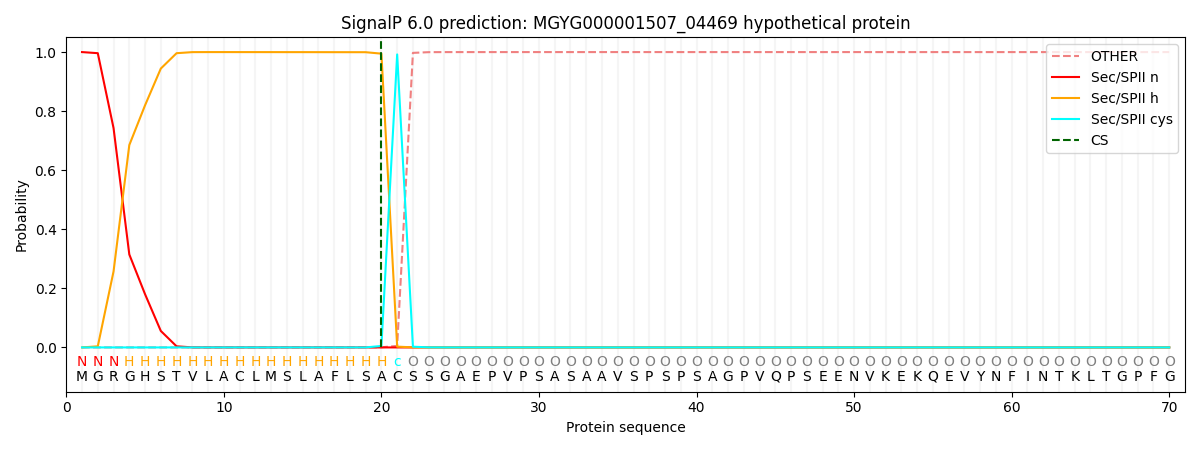
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000045 | 0.000000 | 0.000000 | 0.000000 |
