You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001507_05771
You are here: Home > Sequence: MGYG000001507_05771
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
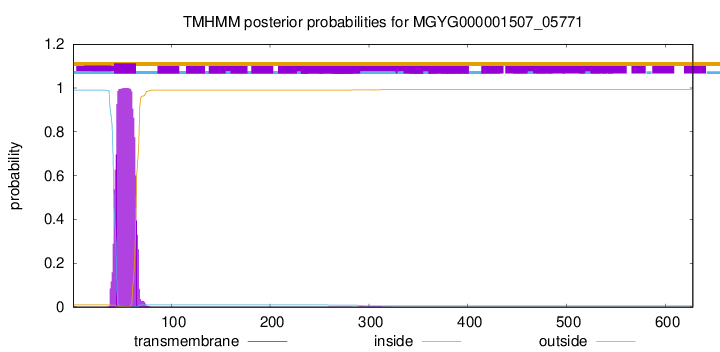
TMHMM annotations
Basic Information help
| Species | Paenibacillus ihuae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus; Paenibacillus ihuae | |||||||||||
| CAZyme ID | MGYG000001507_05771 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 362; End: 2248 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1705 | FlgJ | 7.15e-41 | 323 | 464 | 46 | 189 | Flagellum-specific peptidoglycan hydrolase FlgJ [Cell wall/membrane/envelope biogenesis, Cell motility]. |
| PRK05684 | flgJ | 1.56e-36 | 319 | 454 | 152 | 294 | flagellar assembly peptidoglycan hydrolase FlgJ. |
| COG0739 | NlpD | 1.23e-33 | 8 | 226 | 59 | 264 | Murein DD-endopeptidase MepM and murein hydrolase activator NlpD, contain LysM domain [Cell wall/membrane/envelope biogenesis]. |
| pfam01551 | Peptidase_M23 | 4.65e-30 | 112 | 219 | 1 | 96 | Peptidase family M23. Members of this family are zinc metallopeptidases with a range of specificities. The peptidase family M23 is included in this family, these are Gly-Gly endopeptidases. Peptidase family M23 are also endopeptidases. This family also includes some bacterial lipoproteins for which no proteolytic activity has been demonstrated. This family also includes leukocyte cell-derived chemotaxin 2 (LECT2) proteins. LECT2 is a liver-specific protein which is thought to be linked to hepatocyte growth although the exact function of this protein is unknown. |
| PRK12712 | flgJ | 7.16e-30 | 320 | 454 | 198 | 339 | flagellar rod assembly protein/muramidase FlgJ; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHW35460.1 | 1.15e-270 | 1 | 628 | 1 | 646 |
| AYB48185.1 | 2.15e-176 | 45 | 628 | 27 | 592 |
| AWP25333.1 | 8.36e-176 | 45 | 628 | 27 | 591 |
| AGA60005.1 | 2.99e-168 | 45 | 628 | 42 | 592 |
| QQZ64624.1 | 1.06e-164 | 44 | 628 | 36 | 598 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6UE4_A | 3.44e-22 | 86 | 213 | 237 | 354 | ShyAEndopeptidase from Vibrio cholerae (Closed form) [Vibrio cholerae O1 biovar El Tor str. N16961],6UE4_B ShyA Endopeptidase from Vibrio cholerae (Closed form) [Vibrio cholerae O1 biovar El Tor str. N16961] |
| 6U2A_A | 3.56e-22 | 86 | 213 | 234 | 351 | ShyAendopeptidase from Vibrio cholera (open form) [Vibrio cholerae] |
| 5DN5_A | 3.76e-20 | 328 | 461 | 13 | 152 | Structureof a C-terminally truncated glycoside hydrolase domain from Salmonella typhimurium FlgJ [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],5DN5_B Structure of a C-terminally truncated glycoside hydrolase domain from Salmonella typhimurium FlgJ [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2],5DN5_C Structure of a C-terminally truncated glycoside hydrolase domain from Salmonella typhimurium FlgJ [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
| 5DN4_A | 5.50e-20 | 328 | 461 | 13 | 152 | Structureof the glycoside hydrolase domain from Salmonella typhimurium FlgJ [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
| 3VWO_A | 8.48e-19 | 328 | 459 | 10 | 148 | Crystalstructure of peptidoglycan hydrolase mutant from Sphingomonas sp. A1 [Sphingomonas sp. A1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8K9M4 | 1.86e-18 | 103 | 209 | 280 | 375 | Uncharacterized metalloprotease BUsg_310 OS=Buchnera aphidicola subsp. Schizaphis graminum (strain Sg) OX=198804 GN=BUsg_310 PE=3 SV=1 |
| Q9KQ15 | 3.37e-18 | 322 | 462 | 164 | 311 | Peptidoglycan hydrolase FlgJ OS=Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961) OX=243277 GN=flgJ PE=3 SV=2 |
| Q9I4P4 | 3.89e-18 | 322 | 453 | 242 | 381 | Peptidoglycan hydrolase FlgJ OS=Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) OX=208964 GN=flgJ PE=3 SV=1 |
| P15931 | 4.77e-18 | 328 | 461 | 162 | 301 | Peptidoglycan hydrolase FlgJ OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=flgJ PE=1 SV=1 |
| Q9X9J3 | 7.71e-18 | 328 | 453 | 169 | 301 | Peptidoglycan hydrolase FlgJ OS=Vibrio parahaemolyticus serotype O3:K6 (strain RIMD 2210633) OX=223926 GN=flgJ PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
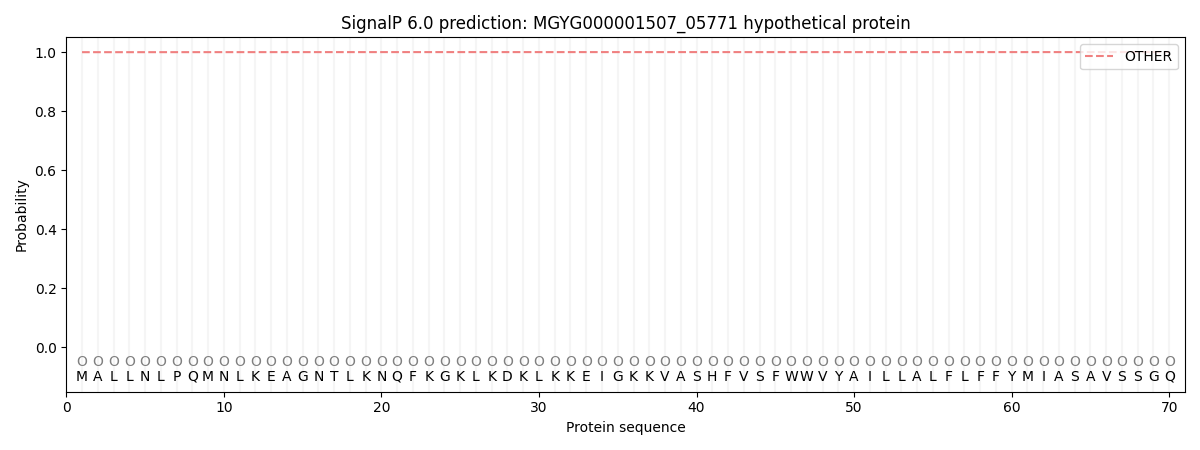
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999874 | 0.000130 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |