You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001509_02180
You are here: Home > Sequence: MGYG000001509_02180
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
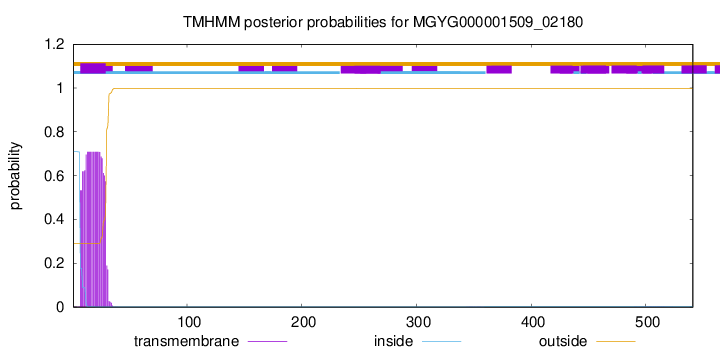
TMHMM annotations
Basic Information help
| Species | Flaviflexus massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; Flaviflexus; Flaviflexus massiliensis | |||||||||||
| CAZyme ID | MGYG000001509_02180 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1405726; End: 1407351 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH23 | 431 | 538 | 3.4e-18 | 0.7925925925925926 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK06347 | PRK06347 | 3.78e-41 | 43 | 297 | 330 | 592 | 1,4-beta-N-acetylmuramoylhydrolase. |
| PRK06347 | PRK06347 | 3.96e-40 | 61 | 366 | 286 | 591 | 1,4-beta-N-acetylmuramoylhydrolase. |
| cd16896 | LT_Slt70-like | 3.38e-24 | 418 | 534 | 5 | 141 | uncharacterized lytic transglycosylase subfamily with similarity to Slt70. Uncharacterized lytic transglycosylase (LT) with a conserved sequence pattern suggesting similarity to the Slt70, a 70kda soluble lytic transglycosylase which also has an N-terminal U-shaped U-domain and a linker L-domain. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| cd00254 | LT-like | 3.23e-22 | 432 | 538 | 1 | 111 | lytic transglycosylase(LT)-like domain. Members include the soluble and insoluble membrane-bound LTs in bacteria and LTs in bacteriophage lambda. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| cd13401 | Slt70-like | 1.42e-16 | 421 | 538 | 10 | 147 | 70kDa soluble lytic transglycosylase (Slt70) and similar proteins. Catalytic domain of the 70kda soluble lytic transglycosylase (LT)-like proteins, which also have an N-terminal U-shaped U-domain and a linker L-domain. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. Proteins similar to this family include the soluble and insoluble membrane-bound LTs in bacteria and the LTs in bacteriophage lambda. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AZN29083.1 | 3.65e-188 | 1 | 541 | 1 | 459 |
| AZQ76365.1 | 2.41e-185 | 1 | 541 | 19 | 485 |
| AXR73717.1 | 3.57e-115 | 114 | 540 | 91 | 543 |
| QCU77827.1 | 2.89e-112 | 113 | 540 | 89 | 502 |
| VEI12524.1 | 2.77e-107 | 113 | 540 | 47 | 494 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2MKX_A | 8.82e-07 | 114 | 159 | 7 | 52 | Solutionstructure of LysM the peptidoglycan binding domain of autolysin AtlA from Enterococcus faecalis [Enterococcus faecalis V583] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q49UX4 | 3.74e-36 | 182 | 378 | 27 | 205 | N-acetylmuramoyl-L-alanine amidase sle1 OS=Staphylococcus saprophyticus subsp. saprophyticus (strain ATCC 15305 / DSM 20229 / NCIMB 8711 / NCTC 7292 / S-41) OX=342451 GN=sle1 PE=3 SV=1 |
| P39046 | 1.15e-33 | 45 | 379 | 256 | 620 | Muramidase-2 OS=Enterococcus hirae (strain ATCC 9790 / DSM 20160 / JCM 8729 / LMG 6399 / NBRC 3181 / NCIMB 6459 / NCDO 1258 / NCTC 12367 / WDCM 00089 / R) OX=768486 GN=EHR_05900 PE=1 SV=1 |
| O31852 | 6.15e-33 | 45 | 300 | 28 | 272 | D-gamma-glutamyl-meso-diaminopimelic acid endopeptidase CwlS OS=Bacillus subtilis (strain 168) OX=224308 GN=cwlS PE=1 SV=1 |
| P37710 | 3.98e-32 | 46 | 378 | 363 | 686 | Autolysin OS=Enterococcus faecalis (strain ATCC 700802 / V583) OX=226185 GN=EF_0799 PE=1 SV=2 |
| O07532 | 3.84e-31 | 43 | 372 | 26 | 356 | Peptidoglycan endopeptidase LytF OS=Bacillus subtilis (strain 168) OX=224308 GN=lytF PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
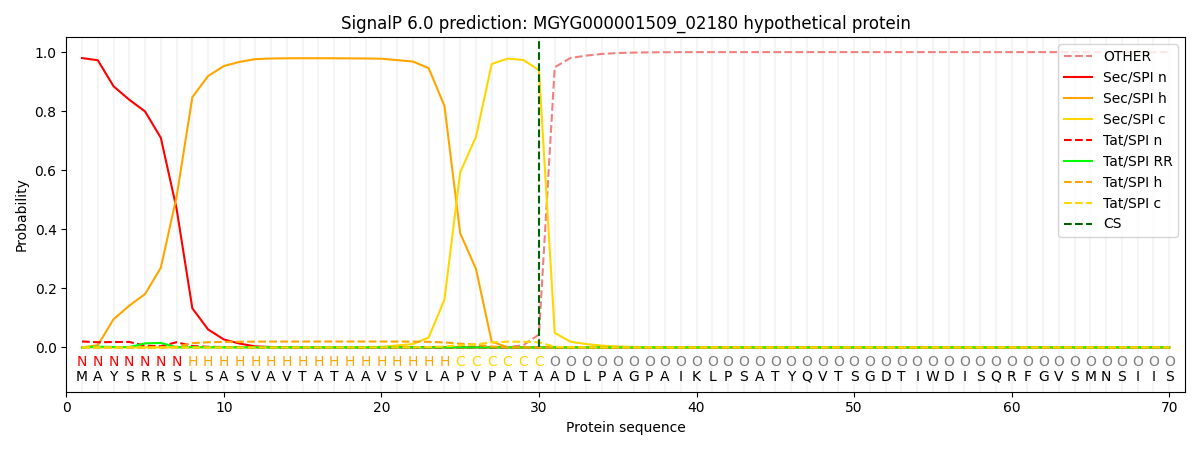
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000823 | 0.977319 | 0.000265 | 0.020953 | 0.000366 | 0.000239 |