You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001510_02419
You are here: Home > Sequence: MGYG000001510_02419
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Numidum massiliense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Thermoactinomycetales; Novibacillaceae; Numidum; Numidum massiliense | |||||||||||
| CAZyme ID | MGYG000001510_02419 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | Trifunctional nucleotide phosphoesterase protein YfkN | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2479023; End: 2480732 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK09419 | PRK09419 | 2.10e-154 | 59 | 568 | 655 | 1152 | multifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase/5'-nucleotidase. |
| COG0737 | UshA | 1.80e-125 | 63 | 564 | 25 | 516 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/5'- or 3'-nucleotidase, 5'-nucleotidase family [Nucleotide transport and metabolism, Defense mechanisms]. |
| PRK09558 | ushA | 6.77e-121 | 59 | 552 | 29 | 538 | bifunctional UDP-sugar hydrolase/5'-nucleotidase periplasmic precursor; Reviewed |
| cd07409 | MPP_CD73_N | 1.91e-103 | 65 | 325 | 1 | 277 | CD73 ecto-5'-nucleotidase and related proteins, N-terminal metallophosphatase domain. CD73 is a mammalian ecto-5'-nucleotidase expressed in endothelial cells and lymphocytes that catalyzes the conversion of 5'-AMP to adenosine in the final step of a pathway that generates adenosine from ATP. This pathway also includes a CD39 nucleoside triphosphate dephosphorylase that mediates the dephosphorylation of ATP to ADP and then to 5'-AMP. These enzymes all have an N-terminal metallophosphatase domain and a C-terminal 5'nucleotidase domain. The N-terminal metallophosphatase domain belongs to a large superfamily of distantly related metallophosphatases (MPPs) that includes: Mre11/SbcD-like exonucleases, Dbr1-like RNA lariat debranching enzymes, YfcE-like phosphodiesterases, purple acid phosphatases (PAPs), YbbF-like UDP-2,3-diacylglucosamine hydrolases, and acid sphingomyelinases (ASMases). MPPs are functionally diverse, but all share a conserved domain with an active site consisting of two metal ions (usually manganese, iron, or zinc) coordinated with octahedral geometry by a cage of histidine, aspartate, and asparagine residues. The conserved domain is a double beta-sheet sandwich with a di-metal active site made up of residues located at the C-terminal side of the sheets. This domain is thought to allow for productive metal coordination. |
| cd00845 | MPP_UshA_N_like | 4.14e-72 | 65 | 327 | 1 | 255 | Escherichia coli UshA-like family, N-terminal metallophosphatase domain. This family includes the bacterial enzyme UshA, and related enzymes including SoxB, CpdB, YhcR, and CD73. All members have a similar domain architecture which includes an N-terminal metallophosphatase domain and a C-terminal nucleotidase domain. The N-terminal metallophosphatase domain belongs to a large superfamily of distantly related metallophosphatases (MPPs) that includes: Mre11/SbcD-like exonucleases, Dbr1-like RNA lariat debranching enzymes, YfcE-like phosphodiesterases, purple acid phosphatases (PAPs), YbbF-like UDP-2,3-diacylglucosamine hydrolases, and acid sphingomyelinases (ASMases). MPPs are functionally diverse, but all share a conserved domain with an active site consisting of two metal ions (usually manganese, iron, or zinc) coordinated with octahedral geometry by a cage of histidine, aspartate, and asparagine residues. The conserved domain is a double beta-sheet sandwich with a di-metal active site made up of residues located at the C-terminal side of the sheets. This domain is thought to allow for productive metal coordination. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QLL63164.1 | 9.04e-114 | 62 | 569 | 25 | 541 |
| QFI67637.1 | 8.67e-113 | 62 | 568 | 25 | 540 |
| CCE97351.1 | 1.36e-112 | 62 | 569 | 25 | 541 |
| AWM26442.1 | 1.36e-112 | 62 | 569 | 25 | 541 |
| APG92489.1 | 1.77e-112 | 62 | 569 | 25 | 541 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2Z1A_A | 1.22e-136 | 61 | 559 | 26 | 528 | Crystalstructure of 5'-nucleotidase precursor from Thermus thermophilus HB8 [Thermus thermophilus HB8] |
| 5H7W_A | 7.95e-91 | 61 | 560 | 1 | 523 | Crystalstructure of 5'-nucleotidase from venom of Naja atra [Naja atra],5H7W_B Crystal structure of 5'-nucleotidase from venom of Naja atra [Naja atra] |
| 7D0V_A | 4.48e-90 | 61 | 560 | 1 | 523 | ChainA, Snake venom 5'-nucleotidase [Naja atra],7D0V_B Chain B, Snake venom 5'-nucleotidase [Naja atra] |
| 6VC9_A | 5.21e-83 | 63 | 559 | 1 | 520 | TB19complex [Homo sapiens],6VCA_A TB38 complex [Homo sapiens],6VCA_B TB38 complex [Homo sapiens],6VCA_C TB38 complex [Homo sapiens],6VCA_D TB38 complex [Homo sapiens] |
| 4H1Y_P | 5.74e-83 | 58 | 559 | 19 | 543 | Humanecto-5'-nucleotidase (CD73): crystal form II (open) in complex with PSB11552 [Homo sapiens],6TVE_P Unliganded human CD73 (5'-nucleotidase) in the open state [Homo sapiens],6TVG_A Human CD73 (ecto 5'-nucleotidase) in complex with AMPCP in the open state [Homo sapiens],7BBJ_A Chain A, 5'-nucleotidase [Homo sapiens],7BBJ_B Chain B, 5'-nucleotidase [Homo sapiens],7P9N_A Chain A, 5'-nucleotidase [Homo sapiens],7P9R_A Chain A, 5'-nucleotidase [Homo sapiens],7P9T_A Chain A, 5'-nucleotidase [Homo sapiens],7PA4_A Chain A, 5'-nucleotidase [Homo sapiens],7PB5_A Chain A, 5'-nucleotidase [Homo sapiens],7PBA_A Chain A, 5'-nucleotidase [Homo sapiens],7PBB_A Chain A, 5'-nucleotidase [Homo sapiens],7PBY_A Chain A, 5'-nucleotidase [Homo sapiens],7PCP_A Chain A, 5'-nucleotidase [Homo sapiens],7PD9_A Chain A, 5'-nucleotidase [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O34313 | 7.20e-145 | 51 | 562 | 655 | 1177 | Trifunctional nucleotide phosphoesterase protein YfkN OS=Bacillus subtilis (strain 168) OX=224308 GN=yfkN PE=1 SV=1 |
| B6EWW8 | 9.66e-91 | 61 | 560 | 40 | 562 | Snake venom 5'-nucleotidase OS=Gloydius brevicaudus OX=259325 PE=2 SV=1 |
| P29240 | 1.02e-90 | 63 | 562 | 30 | 549 | 5'-nucleotidase OS=Diplobatis ommata OX=1870830 PE=2 SV=1 |
| F8S0Z7 | 1.36e-90 | 61 | 560 | 40 | 562 | Snake venom 5'-nucleotidase OS=Crotalus adamanteus OX=8729 PE=1 SV=2 |
| A0A2I4HXH5 | 4.36e-90 | 61 | 560 | 1 | 523 | Snake venom 5'-nucleotidase (Fragment) OS=Naja atra OX=8656 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
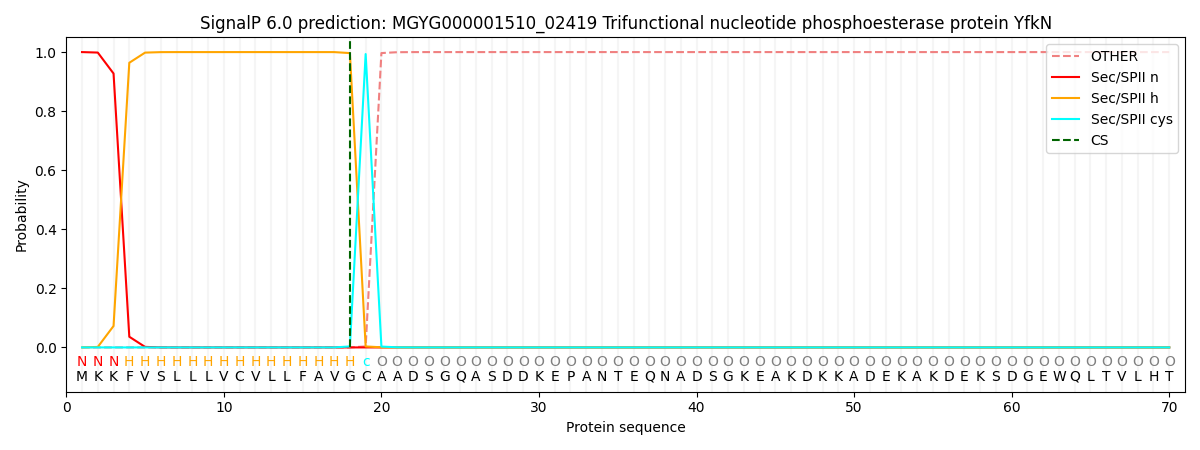
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000054 | 0.000000 | 0.000000 | 0.000000 |
