You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001512_01834
You are here: Home > Sequence: MGYG000001512_01834
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Coprobacter secundus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Coprobacteraceae; Coprobacter; Coprobacter secundus | |||||||||||
| CAZyme ID | MGYG000001512_01834 | |||||||||||
| CAZy Family | PL35 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1072658; End: 1074499 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL35 | 389 | 566 | 4.7e-55 | 0.9776536312849162 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07940 | Hepar_II_III | 1.03e-05 | 385 | 469 | 17 | 90 | Heparinase II/III-like protein. This family features sequences that are similar to a region of the Flavobacterium heparinum proteins heparinase II and heparinase III. The former is known to degrade heparin and heparin sulphate, whereas the latter predominantly degrades heparin sulphate. Both are secreted into the periplasmic space upon induction with heparin. |
| cd04434 | LanC_like | 1.38e-04 | 97 | 260 | 7 | 152 | Cyclases involved in the biosynthesis of lantibiotics, and similar proteins. LanC is the cyclase enzyme of the lanthionine synthetase. Lanthionine is a lantibiotic, a unique class of peptide antibiotics. They are ribosomally synthesized as a precursor peptide and then post-translationally modified to contain thioether cross-links called lanthionines (Lans) or methyllanthionines (MeLans), in addition to 2,3-didehydroalanine (Dha) and (Z)-2,3-didehydrobutyrine (Dhb). These unusual amino acids are introduced by the dehydration of serine and threonine residues, followed by thioether formation via addition of cysteine thiols, catalysed by LanB and LanC or LanM. LanC, the cyclase component, is a zinc metalloprotein, whose bound metal has been proposed to activate the thiol substrate for nucleophilic addition. A related domain is also present in LanM and other pro- and eukaryotic proteins with poorly characterized functions. |
| pfam16332 | DUF4962 | 5.02e-04 | 35 | 220 | 137 | 331 | Domain of unknown function (DUF4962). This family consists of uncharacterized proteins around 870 residues in length and is mainly found in various Bacteroides species. The function of this protein is unknown. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT49750.1 | 9.90e-178 | 25 | 610 | 23 | 605 |
| AYB29714.1 | 5.08e-176 | 15 | 610 | 8 | 599 |
| QNL40059.1 | 7.68e-176 | 6 | 610 | 2 | 601 |
| QGT72630.1 | 1.24e-174 | 6 | 610 | 2 | 601 |
| QUR45369.1 | 7.02e-174 | 6 | 610 | 2 | 601 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
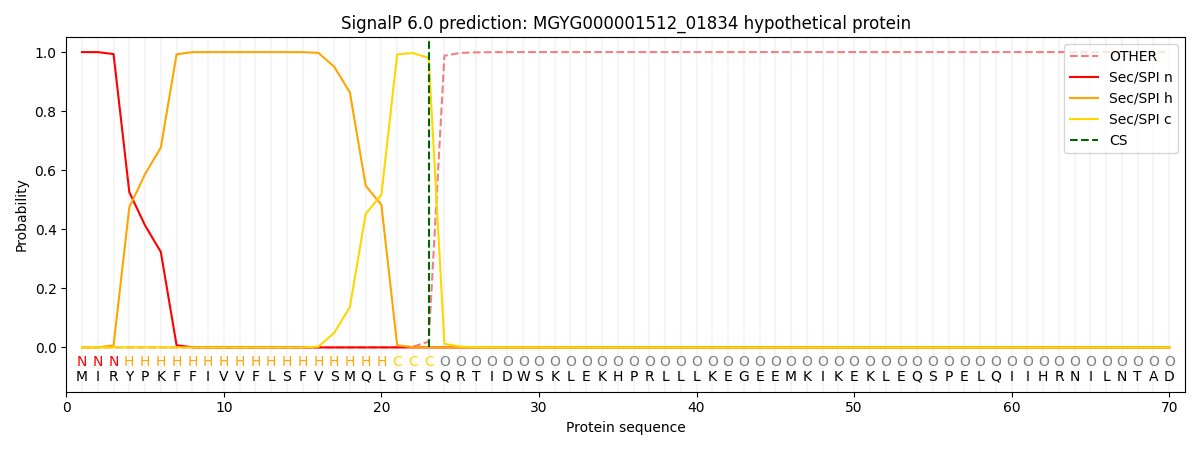
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000215 | 0.999213 | 0.000158 | 0.000137 | 0.000135 | 0.000127 |
