You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001512_01878
You are here: Home > Sequence: MGYG000001512_01878
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Coprobacter secundus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Coprobacteraceae; Coprobacter; Coprobacter secundus | |||||||||||
| CAZyme ID | MGYG000001512_01878 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Endoglucanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1141864; End: 1142916 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 30 | 315 | 8.3e-96 | 0.9858156028368794 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.36e-41 | 42 | 314 | 14 | 271 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 4.23e-28 | 7 | 337 | 16 | 391 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| cd00551 | AmyAc_family | 4.37e-06 | 38 | 176 | 10 | 168 | Alpha amylase catalytic domain family. The Alpha-amylase family comprises the largest family of glycoside hydrolases (GH), with the majority of enzymes acting on starch, glycogen, and related oligo- and polysaccharides. These proteins catalyze the transformation of alpha-1,4 and alpha-1,6 glucosidic linkages with retention of the anomeric center. The protein is described as having 3 domains: A, B, C. A is a (beta/alpha) 8-barrel; B is a loop between the beta 3 strand and alpha 3 helix of A; and C is the C-terminal extension characterized by a Greek key. The majority of the enzymes have an active site cleft found between domains A and B where a triad of catalytic residues (Asp, Glu and Asp) performs catalysis. Other members of this family have lost this catalytic activity as in the case of the human 4F2hc, or only have 2 residues that serve as the catalytic nucleophile and the acid/base, such as Thermus A4 beta-galactosidase with 2 Glu residues (GH42) and human alpha-galactosidase with 2 Asp residues (GH31). The family members are quite extensive and include: alpha amylase, maltosyltransferase, cyclodextrin glycotransferase, maltogenic amylase, neopullulanase, isoamylase, 1,4-alpha-D-glucan maltotetrahydrolase, 4-alpha-glucotransferase, oligo-1,6-glucosidase, amylosucrase, sucrose phosphorylase, and amylomaltase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCI63360.1 | 5.11e-269 | 1 | 350 | 1 | 350 |
| QDH53451.1 | 1.75e-148 | 27 | 339 | 32 | 343 |
| QUT31549.1 | 1.75e-148 | 27 | 339 | 32 | 343 |
| QRM99694.1 | 1.75e-148 | 27 | 339 | 32 | 343 |
| QUT26202.1 | 1.75e-148 | 27 | 339 | 32 | 343 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4EE9_A | 5.25e-93 | 30 | 336 | 6 | 306 | Crystalstructure of the RBcel1 endo-1,4-glucanase [uncultured bacterium],4M24_A Crystal structure of the endo-1,4-glucanase, RBcel1, in complex with cellobiose [uncultured bacterium] |
| 7P6G_A | 1.49e-92 | 30 | 336 | 6 | 306 | ChainA, Endoglucanase [uncultured bacterium],7P6G_B Chain B, Endoglucanase [uncultured bacterium],7P6H_A Chain A, Endoglucanase [uncultured bacterium],7P6H_B Chain B, Endoglucanase [uncultured bacterium] |
| 6ZZ3_A | 2.04e-92 | 30 | 336 | 6 | 306 | ChainA, Endoglucanase [uncultured bacterium],6ZZ3_B Chain B, Endoglucanase [uncultured bacterium],6ZZ3_C Chain C, Endoglucanase [uncultured bacterium],6ZZ3_D Chain D, Endoglucanase [uncultured bacterium] |
| 7P6I_A | 2.11e-92 | 30 | 336 | 6 | 306 | ChainA, Endoglucanase [uncultured bacterium],7P6J_A Chain A, Endoglucanase [uncultured bacterium],7P6J_B Chain B, Endoglucanase [uncultured bacterium],7P6J_C Chain C, Endoglucanase [uncultured bacterium],7P6J_D Chain D, Endoglucanase [uncultured bacterium] |
| 5LJF_A | 4.22e-92 | 30 | 336 | 6 | 306 | ChainA, Endoglucanase [uncultured bacterium],5LJF_B Chain B, Endoglucanase [uncultured bacterium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P58599 | 4.14e-67 | 30 | 337 | 119 | 423 | Endoglucanase OS=Ralstonia solanacearum (strain GMI1000) OX=267608 GN=egl PE=3 SV=1 |
| P17974 | 1.88e-65 | 30 | 337 | 121 | 425 | Endoglucanase OS=Ralstonia solanacearum OX=305 GN=egl PE=1 SV=2 |
| Q2UPQ4 | 3.77e-44 | 27 | 324 | 32 | 318 | Probable endo-beta-1,4-glucanase B OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=eglB PE=3 SV=1 |
| B8MW97 | 3.77e-44 | 27 | 324 | 32 | 318 | probable endo-beta-1,4-glucanase B OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=eglB PE=3 SV=1 |
| Q1HFS8 | 1.37e-42 | 29 | 324 | 30 | 313 | Endo-beta-1,4-glucanase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=eglB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
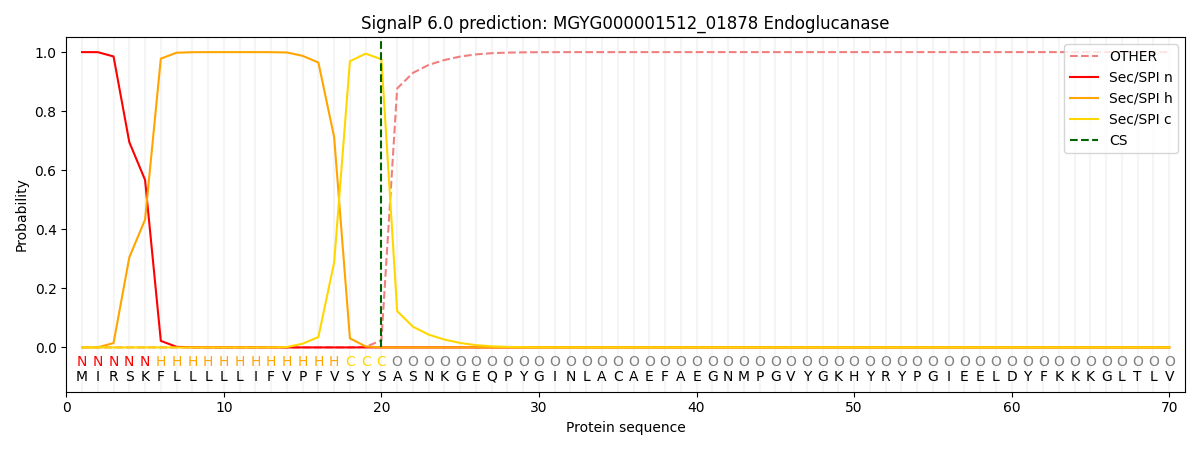
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000344 | 0.998999 | 0.000183 | 0.000156 | 0.000145 | 0.000133 |
