You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001512_01886
You are here: Home > Sequence: MGYG000001512_01886
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
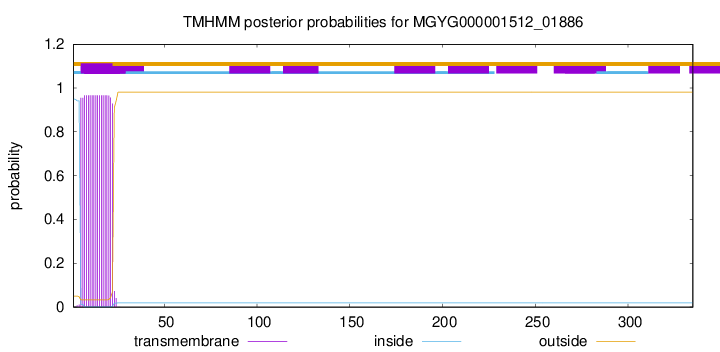
TMHMM annotations
Basic Information help
| Species | Coprobacter secundus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Coprobacteraceae; Coprobacter; Coprobacter secundus | |||||||||||
| CAZyme ID | MGYG000001512_01886 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | Endoglucanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1152188; End: 1153195 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 40 | 315 | 9.3e-120 | 0.996415770609319 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.73e-38 | 41 | 307 | 1 | 271 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 1.42e-14 | 64 | 194 | 67 | 215 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCI63367.1 | 5.44e-261 | 1 | 335 | 1 | 335 |
| AXP79804.1 | 1.10e-128 | 27 | 332 | 23 | 331 |
| QJW90914.1 | 4.84e-125 | 24 | 332 | 19 | 330 |
| QMU30925.1 | 2.24e-124 | 1 | 333 | 1 | 336 |
| AKD02203.1 | 2.97e-124 | 9 | 335 | 9 | 337 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4HTY_A | 3.76e-106 | 18 | 335 | 37 | 359 | CrystalStructure of a metagenome-derived cellulase Cel5A [uncultured bacterium],4HU0_A Crystal Structure of a metagenome-derived cellulase Cel5A in complex with cellotetraose [uncultured bacterium] |
| 5A8N_A | 5.50e-102 | 25 | 331 | 43 | 359 | Crystalstructure of the native form of beta-glucanase SdGluc5_26A from Saccharophagus degradans [Saccharophagus degradans 2-40] |
| 5A8O_A | 1.56e-101 | 25 | 331 | 43 | 359 | Crystalstructure of beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with cellotetraose [Saccharophagus degradans 2-40],5A8P_A Crystal structure beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide B [Saccharophagus degradans 2-40],5A8Q_A Crystal structure beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A obtained by soaking [Saccharophagus degradans 2-40],5A94_A Crystal structure of beta-Glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 1 [Saccharophagus degradans 2-40],5A94_B Crystal structure of beta-Glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 1 [Saccharophagus degradans 2-40],5A94_C Crystal structure of beta-Glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 1 [Saccharophagus degradans 2-40],5A94_D Crystal structure of beta-Glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 1 [Saccharophagus degradans 2-40],5A94_E Crystal structure of beta-Glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 1 [Saccharophagus degradans 2-40],5A94_F Crystal structure of beta-Glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 1 [Saccharophagus degradans 2-40],5A95_A Crystal structure of beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 2 [Saccharophagus degradans 2-40],5A95_B Crystal structure of beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 2 [Saccharophagus degradans 2-40],5A95_C Crystal structure of beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 2 [Saccharophagus degradans 2-40] |
| 5A8M_A | 1.56e-101 | 25 | 330 | 43 | 358 | Crystalstructure of the selenomethionine derivative of beta-glucanase SdGluc5_26A from Saccharophagus degradans [Saccharophagus degradans 2-40],5A8M_B Crystal structure of the selenomethionine derivative of beta-glucanase SdGluc5_26A from Saccharophagus degradans [Saccharophagus degradans 2-40],5A8M_C Crystal structure of the selenomethionine derivative of beta-glucanase SdGluc5_26A from Saccharophagus degradans [Saccharophagus degradans 2-40] |
| 5WH8_A | 3.80e-23 | 33 | 304 | 14 | 273 | CellulaseCel5C_n [uncultured organism] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P22541 | 7.50e-17 | 33 | 302 | 116 | 369 | Endoglucanase A OS=Butyrivibrio fibrisolvens OX=831 GN=celA PE=1 SV=1 |
| P23548 | 1.43e-14 | 2 | 316 | 8 | 367 | Endoglucanase OS=Paenibacillus polymyxa OX=1406 PE=3 SV=2 |
| Q07940 | 4.85e-14 | 76 | 303 | 46 | 257 | Endoglucanase 4 OS=Ruminococcus albus OX=1264 GN=Eg IV PE=1 SV=1 |
| P27035 | 1.88e-13 | 75 | 304 | 200 | 413 | Endoglucanase CelA OS=Streptomyces lividans OX=1916 GN=celA PE=1 SV=2 |
| P06565 | 2.18e-13 | 26 | 302 | 33 | 288 | Endoglucanase B OS=Evansella cellulosilytica (strain ATCC 21833 / DSM 2522 / FERM P-1141 / JCM 9156 / N-4) OX=649639 GN=celB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
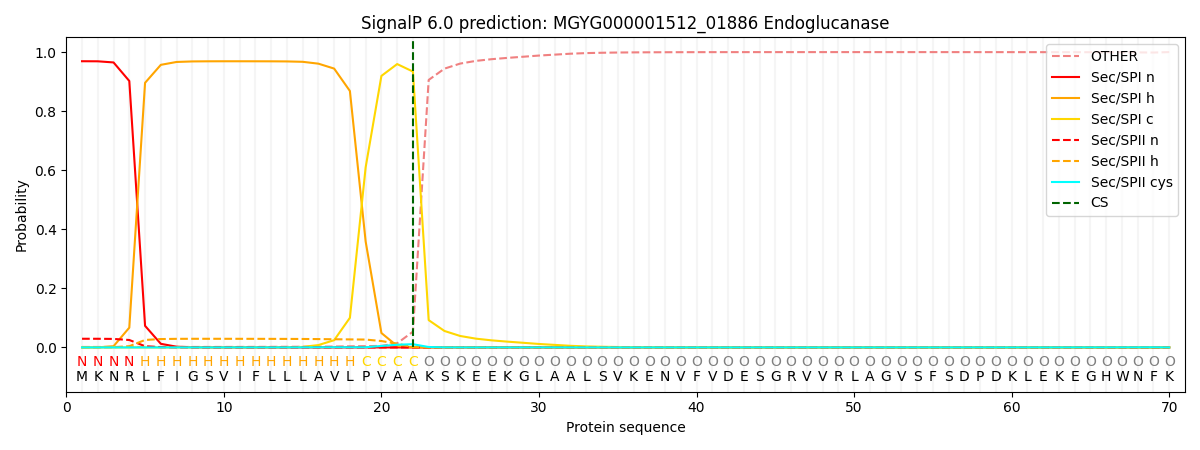
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.004212 | 0.963531 | 0.031426 | 0.000314 | 0.000262 | 0.000244 |