You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001512_01887
You are here: Home > Sequence: MGYG000001512_01887
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
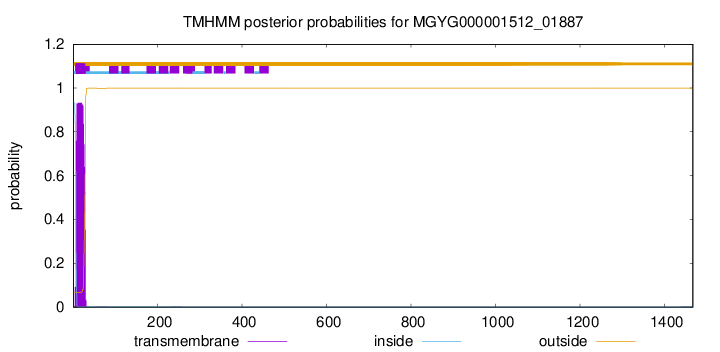
TMHMM annotations
Basic Information help
| Species | Coprobacter secundus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Coprobacteraceae; Coprobacter; Coprobacter secundus | |||||||||||
| CAZyme ID | MGYG000001512_01887 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1153352; End: 1157755 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 1060 | 1341 | 1.3e-79 | 0.975177304964539 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.58e-36 | 1069 | 1339 | 12 | 269 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 1.34e-15 | 1061 | 1361 | 54 | 346 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam18962 | Por_Secre_tail | 9.51e-14 | 1395 | 1465 | 1 | 71 | Secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber have on average twenty or more copies of this C-terminal domain, associated with sorting to the outer membrane and covalent modification. This domain targets proteins to type IX secretion systems and is secreted then cleaved off by a C-terminal signal peptidease. Based on similarity to other families it is likely that this domain adopts an immunoglobulin like fold. |
| TIGR04183 | Por_Secre_tail | 5.28e-11 | 1395 | 1466 | 1 | 71 | Por secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber average twenty or more copies of a C-terminal domain, represented by this model, associated with sorting to the outer membrane and covalent modification. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCI63368.1 | 0.0 | 1 | 1467 | 1 | 1467 |
| QNL39860.1 | 4.50e-94 | 1050 | 1353 | 30 | 341 |
| CBK67191.1 | 5.65e-93 | 1053 | 1353 | 33 | 341 |
| QDM10315.1 | 5.65e-93 | 1053 | 1353 | 33 | 341 |
| QGT72466.1 | 5.65e-93 | 1053 | 1353 | 33 | 341 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4EE9_A | 3.00e-67 | 1059 | 1356 | 4 | 296 | Crystalstructure of the RBcel1 endo-1,4-glucanase [uncultured bacterium],4M24_A Crystal structure of the endo-1,4-glucanase, RBcel1, in complex with cellobiose [uncultured bacterium] |
| 7P6G_A | 7.57e-67 | 1059 | 1356 | 4 | 296 | ChainA, Endoglucanase [uncultured bacterium],7P6G_B Chain B, Endoglucanase [uncultured bacterium],7P6H_A Chain A, Endoglucanase [uncultured bacterium],7P6H_B Chain B, Endoglucanase [uncultured bacterium] |
| 6ZZ3_A | 9.99e-67 | 1059 | 1356 | 4 | 296 | ChainA, Endoglucanase [uncultured bacterium],6ZZ3_B Chain B, Endoglucanase [uncultured bacterium],6ZZ3_C Chain C, Endoglucanase [uncultured bacterium],6ZZ3_D Chain D, Endoglucanase [uncultured bacterium] |
| 7P6I_A | 1.03e-66 | 1059 | 1356 | 4 | 296 | ChainA, Endoglucanase [uncultured bacterium],7P6J_A Chain A, Endoglucanase [uncultured bacterium],7P6J_B Chain B, Endoglucanase [uncultured bacterium],7P6J_C Chain C, Endoglucanase [uncultured bacterium],7P6J_D Chain D, Endoglucanase [uncultured bacterium] |
| 5LJF_A | 1.91e-66 | 1059 | 1356 | 4 | 296 | ChainA, Endoglucanase [uncultured bacterium],5LJF_B Chain B, Endoglucanase [uncultured bacterium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P58599 | 1.44e-38 | 1061 | 1336 | 119 | 389 | Endoglucanase OS=Ralstonia solanacearum (strain GMI1000) OX=267608 GN=egl PE=3 SV=1 |
| P17974 | 5.01e-38 | 1061 | 1336 | 121 | 391 | Endoglucanase OS=Ralstonia solanacearum OX=305 GN=egl PE=1 SV=2 |
| Q1HFS8 | 2.40e-34 | 1061 | 1351 | 31 | 310 | Endo-beta-1,4-glucanase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=eglB PE=1 SV=1 |
| Q5BDU5 | 1.35e-31 | 1060 | 1379 | 24 | 326 | Endo-beta-1,4-glucanase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=eglA PE=1 SV=1 |
| B8MW97 | 2.14e-31 | 1060 | 1353 | 34 | 317 | probable endo-beta-1,4-glucanase B OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=eglB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
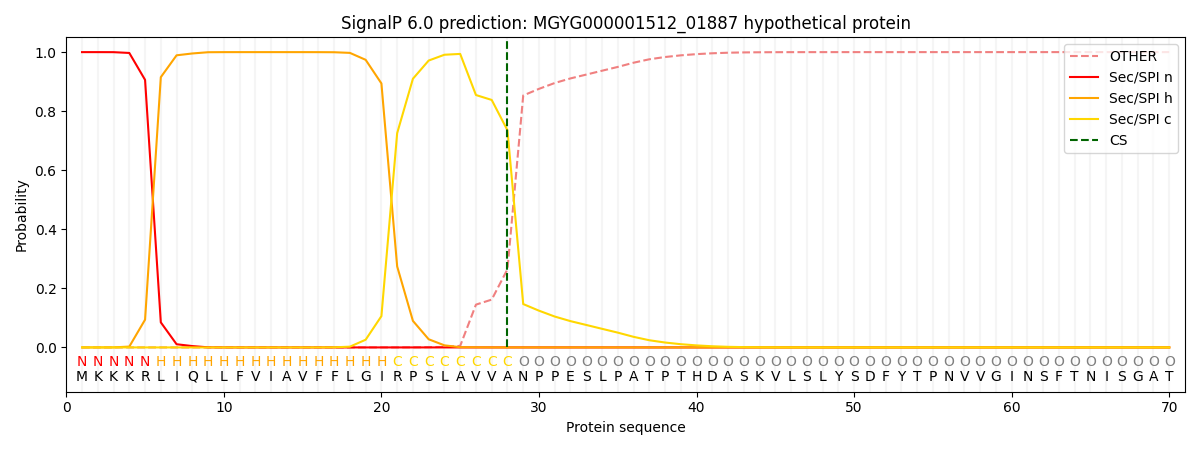
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000364 | 0.998855 | 0.000288 | 0.000158 | 0.000148 | 0.000141 |