You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001512_02863
You are here: Home > Sequence: MGYG000001512_02863
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
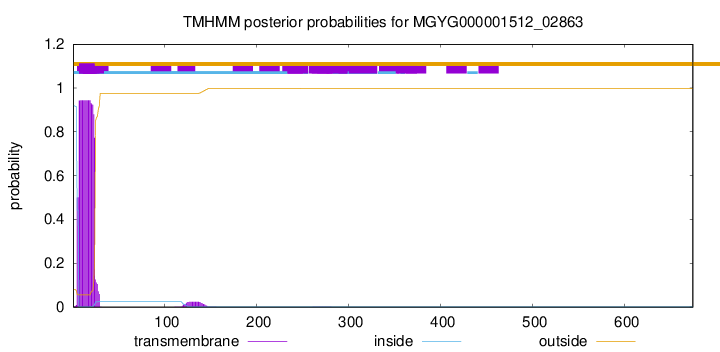
TMHMM annotations
Basic Information help
| Species | Coprobacter secundus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Coprobacteraceae; Coprobacter; Coprobacter secundus | |||||||||||
| CAZyme ID | MGYG000001512_02863 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1085168; End: 1087192 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM13 | 439 | 602 | 3.7e-16 | 0.7925531914893617 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08757 | CotH | 3.59e-33 | 73 | 409 | 1 | 311 | CotH kinase protein. Members of this family include the spore coat protein H (cotH). This protein is an atypical protein kinase that phosphorylates CotB and CotG. |
| pfam14200 | RicinB_lectin_2 | 2.48e-10 | 434 | 497 | 7 | 73 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 3.10e-08 | 474 | 550 | 2 | 73 | Ricin-type beta-trefoil lectin domain-like. |
| pfam00652 | Ricin_B_lectin | 6.69e-04 | 441 | 550 | 2 | 100 | Ricin-type beta-trefoil lectin domain. |
| TIGR04183 | Por_Secre_tail | 0.002 | 628 | 674 | 24 | 72 | Por secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber average twenty or more copies of a C-terminal domain, represented by this model, associated with sorting to the outer membrane and covalent modification. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCI64517.1 | 0.0 | 1 | 674 | 1 | 674 |
| QRR03748.1 | 1.47e-111 | 1 | 586 | 1 | 566 |
| CUH93331.1 | 1.03e-48 | 21 | 425 | 235 | 704 |
| APF19010.1 | 2.89e-36 | 24 | 422 | 23 | 456 |
| SCG56726.1 | 1.78e-34 | 23 | 383 | 145 | 525 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
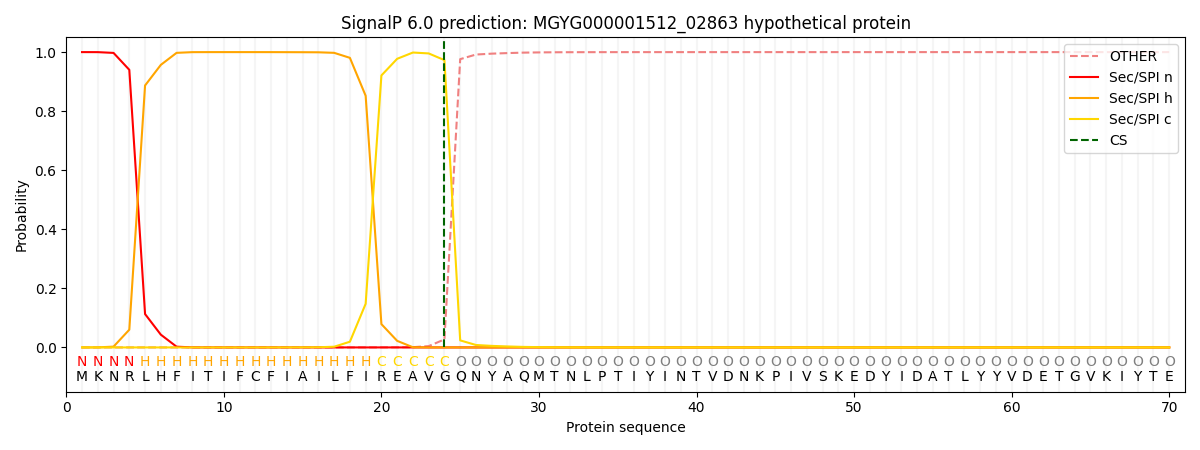
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000451 | 0.998754 | 0.000252 | 0.000175 | 0.000171 | 0.000161 |