You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001512_03070
You are here: Home > Sequence: MGYG000001512_03070
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
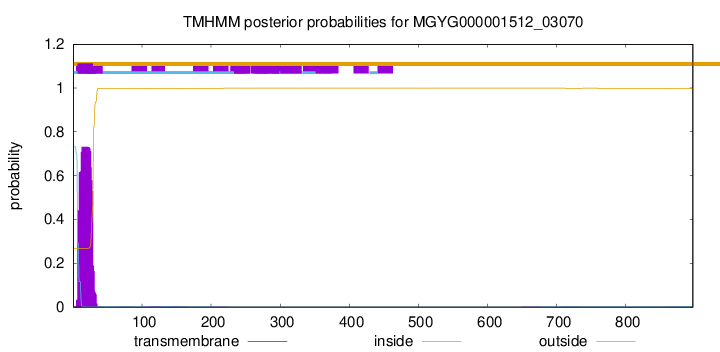
TMHMM annotations
Basic Information help
| Species | Coprobacter secundus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Coprobacteraceae; Coprobacter; Coprobacter secundus | |||||||||||
| CAZyme ID | MGYG000001512_03070 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1330027; End: 1332720 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 411 | 748 | 1.9e-66 | 0.9323076923076923 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 1.20e-32 | 393 | 628 | 89 | 366 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| PLN02188 | PLN02188 | 1.58e-07 | 389 | 569 | 39 | 218 | polygalacturonase/glycoside hydrolase family protein |
| TIGR04183 | Por_Secre_tail | 5.13e-06 | 831 | 895 | 6 | 70 | Por secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber average twenty or more copies of a C-terminal domain, represented by this model, associated with sorting to the outer membrane and covalent modification. |
| PLN02793 | PLN02793 | 3.43e-05 | 381 | 571 | 47 | 242 | Probable polygalacturonase |
| PLN03003 | PLN03003 | 2.88e-04 | 478 | 623 | 112 | 283 | Probable polygalacturonase At3g15720 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCI64737.1 | 0.0 | 1 | 897 | 1 | 897 |
| QUT74982.1 | 0.0 | 3 | 894 | 1 | 895 |
| QUT74984.1 | 2.75e-107 | 389 | 812 | 25 | 450 |
| ALJ60390.1 | 1.09e-104 | 393 | 815 | 31 | 456 |
| QUT88603.1 | 2.98e-104 | 393 | 815 | 31 | 456 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5OLP_A | 1.03e-24 | 393 | 626 | 51 | 325 | Galacturonidase[Bacteroides thetaiotaomicron VPI-5482],5OLP_B Galacturonidase [Bacteroides thetaiotaomicron VPI-5482] |
| 3JUR_A | 1.05e-16 | 385 | 631 | 27 | 325 | Thecrystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_B The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_C The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_D The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima] |
| 4MXN_A | 2.29e-11 | 393 | 605 | 28 | 235 | Crystalstructure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_B Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_C Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_D Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184] |
| 2UVE_A | 9.23e-09 | 389 | 581 | 159 | 405 | Structureof Yersinia enterocolitica Family 28 Exopolygalacturonase [Yersinia enterocolitica],2UVE_B Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase [Yersinia enterocolitica],2UVF_A Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase in Complex with Digalaturonic Acid [Yersinia enterocolitica],2UVF_B Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase in Complex with Digalaturonic Acid [Yersinia enterocolitica] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0DJ97 | 4.26e-12 | 29 | 154 | 35 | 161 | Putative Gly-rich membrane protein Bcell_0380 OS=Evansella cellulosilytica (strain ATCC 21833 / DSM 2522 / FERM P-1141 / JCM 9156 / N-4) OX=649639 GN=Bcell_0380 PE=4 SV=1 |
| Q9FY19 | 1.40e-11 | 389 | 756 | 62 | 415 | Polygalacturonase OS=Juniperus ashei OX=13101 GN=JNA2 PE=1 SV=1 |
| P15922 | 2.43e-09 | 393 | 581 | 158 | 398 | Exo-poly-alpha-D-galacturonosidase OS=Dickeya chrysanthemi OX=556 GN=pehX PE=1 SV=1 |
| Q18DN4 | 9.38e-09 | 28 | 172 | 1579 | 1723 | Halomucin OS=Haloquadratum walsbyi (strain DSM 16790 / HBSQ001) OX=362976 GN=hmu PE=3 SV=1 |
| Q7M1E7 | 5.87e-08 | 389 | 619 | 61 | 291 | Polygalacturonase OS=Chamaecyparis obtusa OX=13415 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
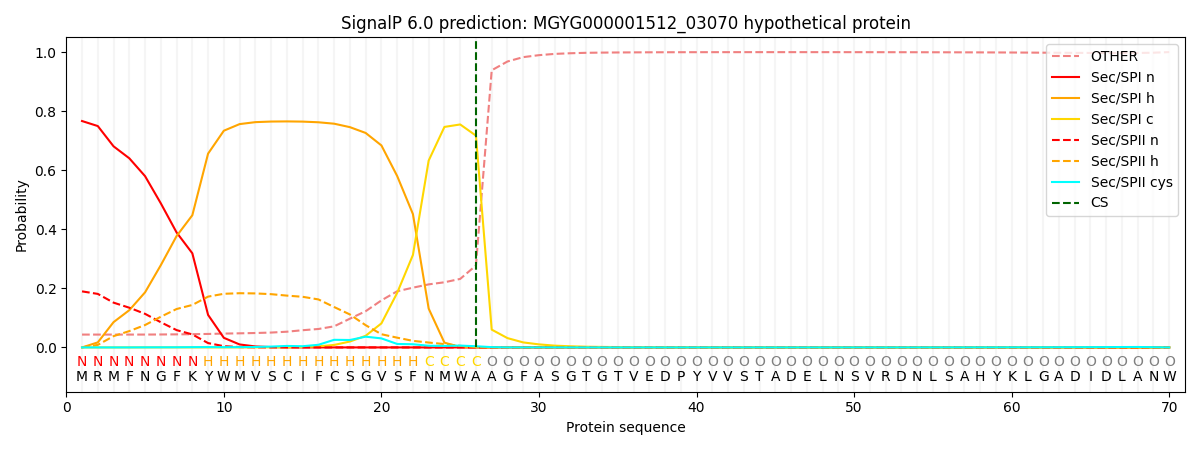
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.050647 | 0.751541 | 0.196309 | 0.000575 | 0.000384 | 0.000529 |