You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001513_00746
You are here: Home > Sequence: MGYG000001513_00746
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Clostridium_P massiliamazoniense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium_P; Clostridium_P massiliamazoniense | |||||||||||
| CAZyme ID | MGYG000001513_00746 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 572965; End: 574632 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08239 | SH3_3 | 6.95e-13 | 430 | 481 | 1 | 54 | Bacterial SH3 domain. |
| pfam08239 | SH3_3 | 7.32e-12 | 506 | 554 | 3 | 53 | Bacterial SH3 domain. |
| COG3103 | YgiM | 1.83e-11 | 432 | 554 | 34 | 149 | Uncharacterized conserved protein YgiM, contains N-terminal SH3 domain, DUF1202 family [General function prediction only]. |
| pfam08239 | SH3_3 | 4.76e-10 | 333 | 388 | 1 | 54 | Bacterial SH3 domain. |
| NF038016 | sporang_Gsm | 5.65e-10 | 432 | 555 | 1 | 141 | sporangiospore maturation cell wall hydrolase GsmA. The peptidoglycan-hydrolyzing enzyme GsmA occurs in some sporangia-forming members of the Actinobacteria, such as Actinoplanes missouriensis, and is required for proper separation of spores. GsmA proteins have one or two SH3 domains N-terminal to the hydrolase domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QLY80841.1 | 1.17e-106 | 108 | 554 | 396 | 849 |
| QPJ84712.1 | 6.13e-100 | 105 | 482 | 64 | 413 |
| QAA30551.1 | 1.56e-76 | 107 | 555 | 44 | 536 |
| AXH52446.1 | 3.26e-46 | 315 | 555 | 715 | 949 |
| QQA11114.1 | 3.15e-43 | 328 | 554 | 473 | 693 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2KRS_A | 5.19e-15 | 500 | 554 | 5 | 59 | SolutionNMR structure of SH3 domain from CPF_0587 (fragment 415-479) from Clostridium perfringens. Northeast Structural Genomics Consortium (NESG) Target CpR74A. [Clostridium perfringens] |
| 2KT8_A | 4.87e-14 | 419 | 487 | 1 | 68 | SolutionNMR structure of the CPE1231(468-535) protein from Clostridium perfringens, Northeast Structural Genomics Consortium Target CpR82B [Clostridium perfringens] |
| 2KYB_A | 4.73e-12 | 425 | 480 | 5 | 60 | Solutionstructure of CpR82G from Clostridium perfringens. North East Structural Genomics Consortium Target CpR82g [Clostridium perfringens ATCC 13124] |
| 2KQ8_A | 6.14e-12 | 422 | 484 | 2 | 64 | ChainA, Cell wall hydrolase [[Bacillus thuringiensis] serovar konkukian] |
| 4KRT_A | 4.51e-10 | 427 | 555 | 227 | 351 | X-raystructure of endolysin from clostridium perfringens phage phiSM101 [Clostridium phage phiSM101],4KRT_B X-ray structure of endolysin from clostridium perfringens phage phiSM101 [Clostridium phage phiSM101] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O51481 | 1.24e-20 | 114 | 274 | 29 | 202 | Uncharacterized protein BB_0531 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0531 PE=3 SV=1 |
| O32041 | 5.88e-16 | 326 | 554 | 30 | 239 | Putative N-acetylmuramoyl-L-alanine amidase YrvJ OS=Bacillus subtilis (strain 168) OX=224308 GN=yrvJ PE=3 SV=1 |
| P08696 | 6.68e-08 | 423 | 484 | 575 | 636 | Bacteriocin BCN5 OS=Clostridium perfringens OX=1502 GN=bcn PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
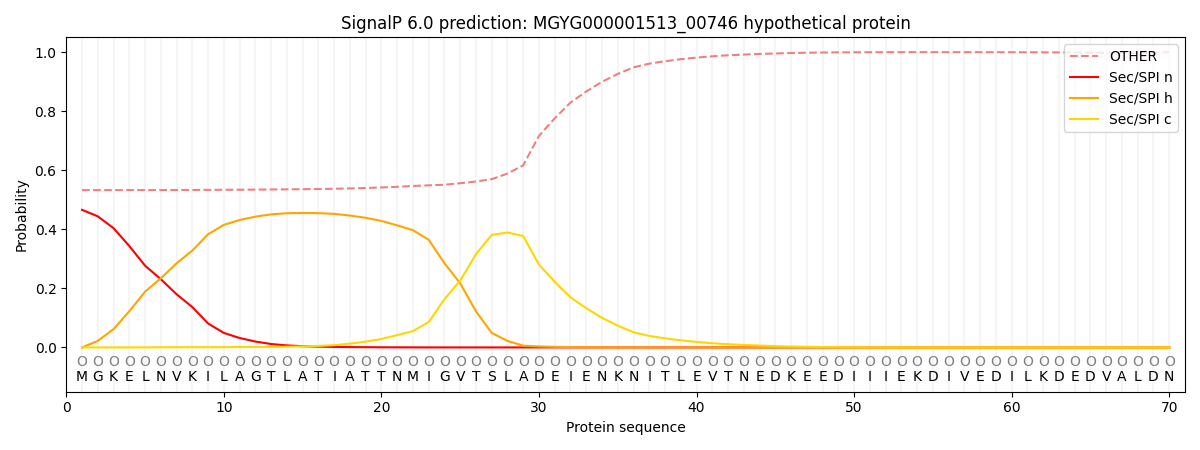
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.555633 | 0.441224 | 0.001819 | 0.000650 | 0.000302 | 0.000367 |
