You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001514_00040
You are here: Home > Sequence: MGYG000001514_00040
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
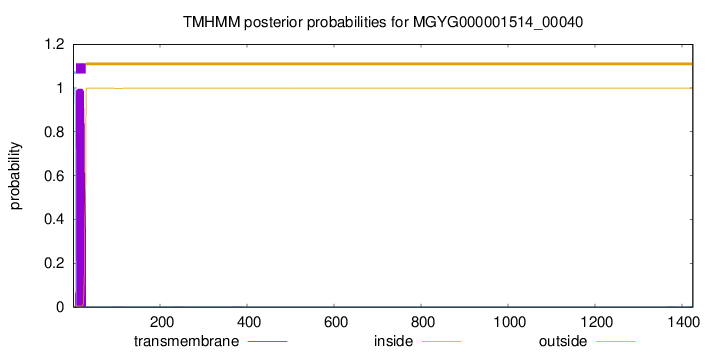
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A ihumii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A ihumii | |||||||||||
| CAZyme ID | MGYG000001514_00040 | |||||||||||
| CAZy Family | CBM54 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 42186; End: 46463 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 431 | 747 | 1.1e-66 | 0.9141914191419142 |
| CBM23 | 1080 | 1233 | 1.9e-46 | 0.9259259259259259 |
| CBM59 | 1280 | 1423 | 2.4e-29 | 0.9793103448275862 |
| CBM54 | 195 | 306 | 3.7e-27 | 0.9736842105263158 |
| CBM27 | 896 | 1066 | 1.4e-24 | 0.9940476190476191 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02156 | Glyco_hydro_26 | 6.78e-55 | 431 | 699 | 1 | 263 | Glycosyl hydrolase family 26. |
| COG4124 | ManB2 | 4.48e-20 | 519 | 707 | 103 | 297 | Beta-mannanase [Carbohydrate transport and metabolism]. |
| pfam09212 | CBM27 | 2.01e-12 | 894 | 1066 | 7 | 170 | Carbohydrate binding module 27. Members of this family are carbohydrate binding modules that bind to beta-1, 4-manno-oligosaccharides, carob galactomannan, and konjac glucomannan, but not to cellulose (insoluble and soluble) or soluble birchwood xylan. They adopt a beta sandwich structure comprising 13 beta strands with a single, small alpha-helix and a single metal atom. |
| pfam03425 | CBM_11 | 1.60e-11 | 1079 | 1195 | 6 | 129 | Carbohydrate binding domain (family 11). |
| NF033190 | inl_like_NEAT_1 | 4.41e-08 | 35 | 191 | 589 | 743 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AIQ63112.1 | 0.0 | 1 | 1423 | 1 | 1424 |
| QMV42451.1 | 0.0 | 8 | 1425 | 5 | 1397 |
| ANY66447.1 | 0.0 | 2 | 1424 | 3 | 1400 |
| AWV31354.1 | 0.0 | 1 | 1422 | 1 | 1260 |
| ADL67722.1 | 0.0 | 7 | 1425 | 8 | 1409 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4YN5_A | 7.36e-134 | 430 | 808 | 51 | 442 | Catalyticdomain of Bacillus sp. JAMB-750 GH26 Endo-beta-1,4-mannanase [Bacillus sp. JAMB750] |
| 2BVT_A | 6.00e-121 | 428 | 871 | 6 | 448 | Thestructure of a modular endo-beta-1,4-mannanase from Cellulomonas fimi explains the product specificity of glycoside hydrolase family 26 mannanases. [Cellulomonas fimi],2BVT_B The structure of a modular endo-beta-1,4-mannanase from Cellulomonas fimi explains the product specificity of glycoside hydrolase family 26 mannanases. [Cellulomonas fimi],2BVY_A The structure and characterization of a modular endo-beta-1,4-mannanase from Cellulomonas fimi [Cellulomonas fimi] |
| 2X2Y_A | 4.21e-119 | 428 | 871 | 6 | 448 | Cellulomonasfimi endo-beta-1,4-mannanase double mutant [Cellulomonas fimi],2X2Y_B Cellulomonas fimi endo-beta-1,4-mannanase double mutant [Cellulomonas fimi] |
| 3TP4_A | 1.67e-109 | 428 | 871 | 6 | 448 | CrystalStructure of engineered protein at the resolution 1.98A, Northeast Structural Genomics Consortium Target OR128 [synthetic construct],3TP4_B Crystal Structure of engineered protein at the resolution 1.98A, Northeast Structural Genomics Consortium Target OR128 [synthetic construct] |
| 1J9Y_A | 2.00e-91 | 428 | 791 | 9 | 379 | Crystalstructure of mannanase 26A from Pseudomonas cellulosa [Cellvibrio japonicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P49424 | 3.54e-90 | 428 | 791 | 47 | 417 | Mannan endo-1,4-beta-mannosidase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=manA PE=1 SV=2 |
| A1A278 | 3.00e-79 | 428 | 1193 | 40 | 834 | Mannan endo-1,4-beta-mannosidase OS=Bifidobacterium adolescentis (strain ATCC 15703 / DSM 20083 / NCTC 11814 / E194a) OX=367928 GN=BAD_1030 PE=1 SV=1 |
| O05512 | 1.11e-17 | 480 | 671 | 81 | 269 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis (strain 168) OX=224308 GN=gmuG PE=1 SV=2 |
| P55278 | 3.51e-17 | 480 | 671 | 79 | 267 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis OX=1423 PE=3 SV=1 |
| C6CRV0 | 8.79e-17 | 24 | 199 | 1279 | 1458 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
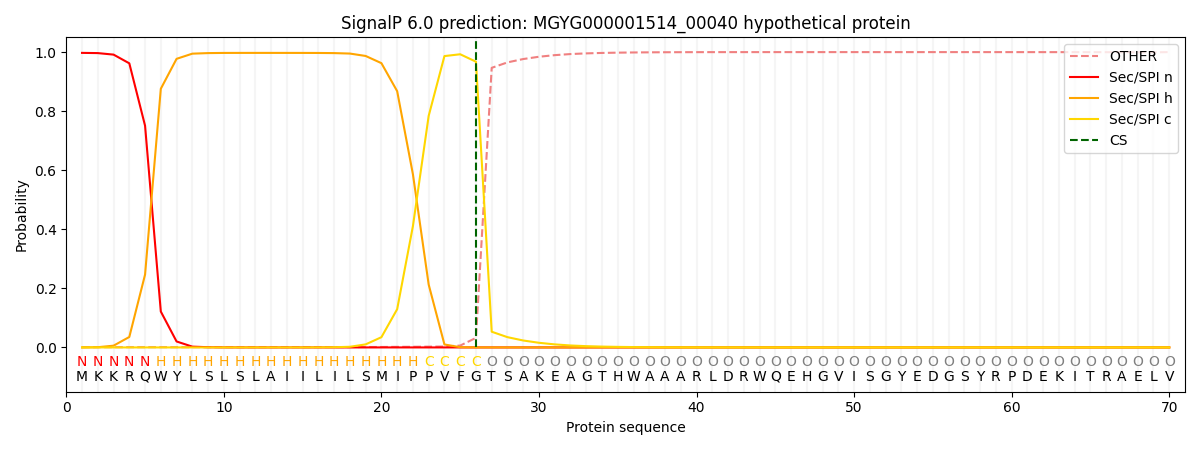
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002666 | 0.993979 | 0.002550 | 0.000313 | 0.000241 | 0.000210 |