You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001514_00608
You are here: Home > Sequence: MGYG000001514_00608
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
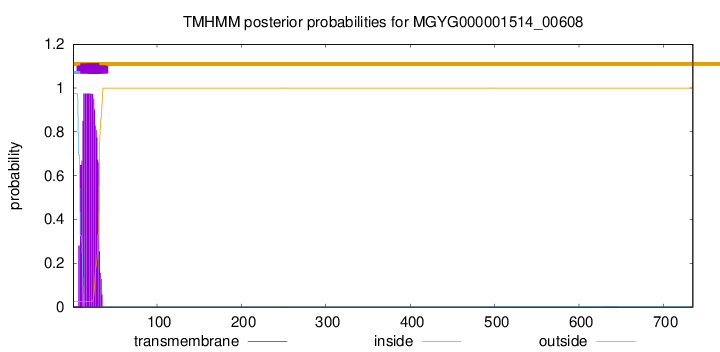
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A ihumii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A ihumii | |||||||||||
| CAZyme ID | MGYG000001514_00608 | |||||||||||
| CAZy Family | CBM22 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 35751; End: 37958 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 228 | 508 | 1.2e-79 | 0.9603960396039604 |
| CBM22 | 40 | 172 | 8.3e-33 | 0.9923664122137404 |
| CBM3 | 590 | 671 | 2.6e-24 | 0.9886363636363636 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 1.49e-73 | 269 | 492 | 2 | 241 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 1.06e-59 | 233 | 493 | 8 | 283 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 4.03e-43 | 230 | 495 | 28 | 313 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00942 | CBM_3 | 4.42e-26 | 589 | 670 | 1 | 82 | Cellulose binding domain. |
| pfam02018 | CBM_4_9 | 4.30e-23 | 39 | 175 | 1 | 134 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AZK46486.1 | 0.0 | 1 | 735 | 2 | 736 |
| ANV74926.1 | 2.41e-202 | 37 | 597 | 27 | 586 |
| ALX07190.1 | 2.41e-202 | 37 | 597 | 27 | 586 |
| ABN53790.1 | 2.41e-202 | 37 | 597 | 27 | 586 |
| ADU73272.1 | 2.41e-202 | 37 | 597 | 27 | 586 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7D88_A | 6.35e-155 | 187 | 570 | 38 | 423 | ChainA, Beta-xylanase [Bacillus sp. (in: Bacteria)] |
| 7D89_A | 4.05e-153 | 187 | 570 | 38 | 423 | ChainA, Beta-xylanase [Bacillus sp. (in: Bacteria)] |
| 6D5B_A | 4.96e-38 | 584 | 735 | 16 | 167 | Structureof Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_B Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_C Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_D Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_E Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_F Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_G Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_H Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_I Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_J Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_K Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5B_L Structure of Caldicellulosiruptor danielii CBM3 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii] |
| 3WUF_A | 5.55e-38 | 237 | 498 | 16 | 293 | Themutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) from Streptomyces sp. 9 [Streptomyces sp.],3WUG_A The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) with xylobiose from Streptomyces sp. 9 [Streptomyces sp.] |
| 3WUB_A | 1.93e-37 | 237 | 498 | 16 | 293 | Thewild type crystal structure of b-1,4-Xylanase (XynAS9) from Streptomyces sp. 9 [Streptomyces sp.],3WUE_A The wild type crystal structure of b-1,4-Xylanase (XynAS9) with xylobiose from Streptomyces sp. 9 [Streptomyces sp.] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A3DH97 | 7.06e-142 | 191 | 572 | 378 | 759 | Anti-sigma-I factor RsgI6 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=rsgI6 PE=1 SV=1 |
| A0A1P8AWH8 | 5.12e-106 | 40 | 571 | 397 | 943 | Endo-1,4-beta-xylanase 1 OS=Arabidopsis thaliana OX=3702 GN=XYN1 PE=1 SV=1 |
| F4JG10 | 1.92e-101 | 79 | 572 | 250 | 749 | Endo-1,4-beta-xylanase 3 OS=Arabidopsis thaliana OX=3702 GN=XYN3 PE=2 SV=1 |
| O80596 | 1.15e-98 | 28 | 572 | 505 | 1063 | Endo-1,4-beta-xylanase 2 OS=Arabidopsis thaliana OX=3702 GN=XYN2 PE=3 SV=1 |
| Q84WT5 | 8.76e-69 | 97 | 547 | 83 | 539 | Endo-1,4-beta-xylanase 5-like OS=Arabidopsis thaliana OX=3702 GN=At4g33820 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
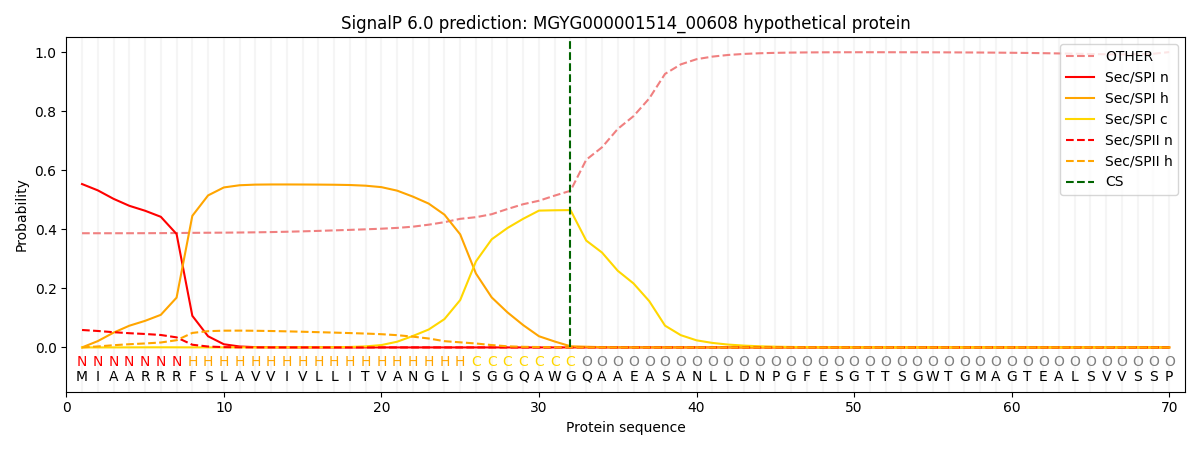
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.402342 | 0.530256 | 0.062144 | 0.002274 | 0.001025 | 0.001940 |