You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001514_00609
You are here: Home > Sequence: MGYG000001514_00609
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A ihumii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A ihumii | |||||||||||
| CAZyme ID | MGYG000001514_00609 | |||||||||||
| CAZy Family | GH6 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 38053; End: 40233 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH6 | 62 | 411 | 6.6e-95 | 0.9455782312925171 |
| CBM3 | 581 | 661 | 1.1e-26 | 0.9772727272727273 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5297 | CelA1 | 0.0 | 34 | 466 | 18 | 446 | Cellulase/cellobiase CelA1 [Carbohydrate transport and metabolism]. |
| pfam01341 | Glyco_hydro_6 | 6.37e-130 | 66 | 413 | 1 | 292 | Glycosyl hydrolases family 6. |
| pfam00942 | CBM_3 | 1.12e-24 | 580 | 661 | 1 | 82 | Cellulose binding domain. |
| smart01067 | CBM_3 | 1.01e-20 | 580 | 661 | 1 | 82 | Cellulose binding domain. |
| cd00063 | FN3 | 1.94e-05 | 491 | 563 | 13 | 91 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AZK46485.1 | 0.0 | 1 | 726 | 1 | 725 |
| AZS14930.1 | 0.0 | 7 | 726 | 7 | 809 |
| AIQ36735.1 | 2.13e-311 | 1 | 726 | 1 | 804 |
| AUS25285.1 | 8.72e-308 | 3 | 726 | 4 | 714 |
| QDA25983.1 | 1.12e-306 | 3 | 726 | 7 | 717 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5XYH_A | 5.13e-207 | 35 | 463 | 1 | 425 | CrystalStructure of catalytic domain of 1,4-beta-Cellobiosidase (CbsA) from Xanthomonas oryzae pv. oryzae [Xanthomonas oryzae pv. oryzae] |
| 7CBD_A | 3.79e-176 | 34 | 474 | 2 | 438 | Catalyticdomain of Cellulomonas fimi Cel6B [Cellulomonas fimi ATCC 484] |
| 6K55_A | 1.30e-160 | 38 | 463 | 2 | 426 | Inactivatedmutant (D140A) of Hyperthermophilic GH6 cellobiohydrolase II (HmCel6A) in complex with hexasaccharide [Ardenticatenia bacterium],6K55_B Inactivated mutant (D140A) of Hyperthermophilic GH6 cellobiohydrolase II (HmCel6A) in complex with hexasaccharide [Ardenticatenia bacterium],6K55_C Inactivated mutant (D140A) of Hyperthermophilic GH6 cellobiohydrolase II (HmCel6A) in complex with hexasaccharide [Ardenticatenia bacterium] |
| 4B4F_A | 3.24e-126 | 38 | 463 | 3 | 420 | Thermobifidafusca Cel6B(E3) co-crystallized with cellobiose [Thermobifida fusca],4B4F_B Thermobifida fusca Cel6B(E3) co-crystallized with cellobiose [Thermobifida fusca],4B4H_A Thermobifida fusca cellobiohydrolase Cel6B(E3) catalytic domain [Thermobifida fusca],4B4H_B Thermobifida fusca cellobiohydrolase Cel6B(E3) catalytic domain [Thermobifida fusca] |
| 4AVO_A | 5.08e-125 | 38 | 463 | 3 | 420 | Thermobifidafusca cellobiohydrolase Cel6B catalytic mutant D274A cocrystallized with cellobiose [Thermobifida fusca YX] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P50401 | 1.64e-172 | 30 | 488 | 37 | 490 | Exoglucanase A OS=Cellulomonas fimi (strain ATCC 484 / DSM 20113 / JCM 1341 / NBRC 15513 / NCIMB 8980 / NCTC 7547) OX=590998 GN=cbhA PE=1 SV=1 |
| A1CCN4 | 7.20e-57 | 40 | 463 | 108 | 464 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=cbhC PE=3 SV=1 |
| P49075 | 1.43e-56 | 40 | 463 | 90 | 438 | Exoglucanase 3 OS=Agaricus bisporus OX=5341 GN=cel3 PE=1 SV=1 |
| Q5B2E8 | 7.79e-56 | 40 | 463 | 96 | 455 | 1,4-beta-D-glucan cellobiohydrolase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=cbhC PE=1 SV=2 |
| A2QYR9 | 1.13e-54 | 24 | 463 | 84 | 459 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=cbhC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
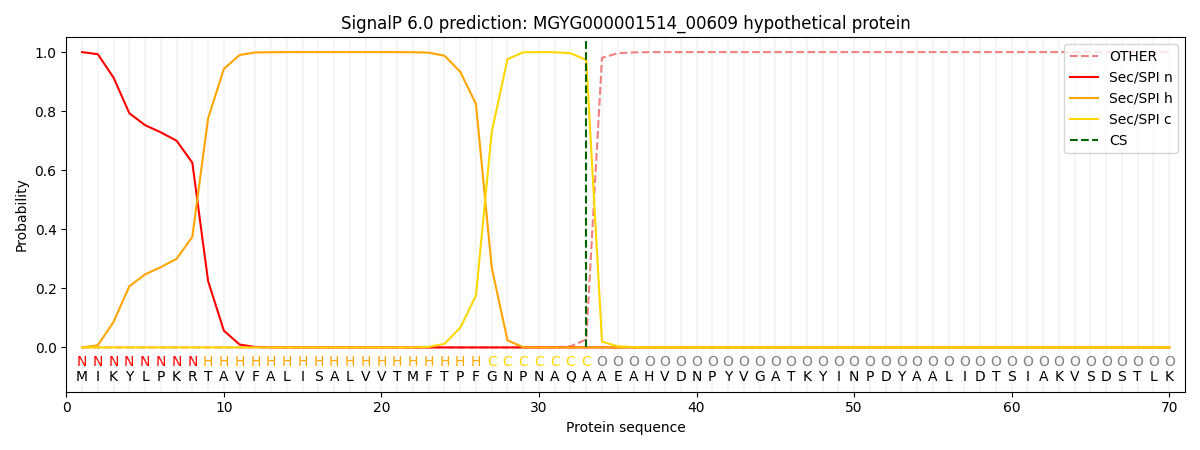
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000311 | 0.998906 | 0.000193 | 0.000219 | 0.000188 | 0.000161 |
