You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001514_04050
You are here: Home > Sequence: MGYG000001514_04050
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
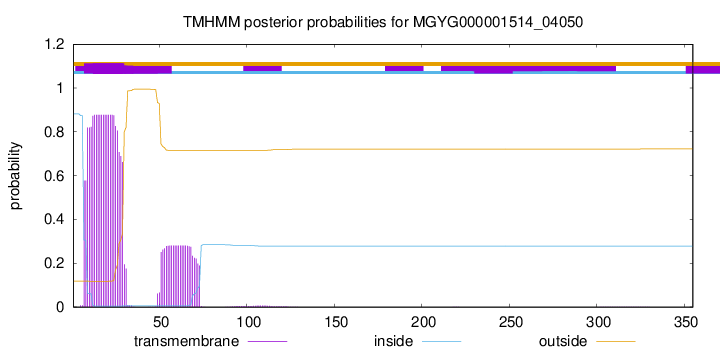
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A ihumii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A ihumii | |||||||||||
| CAZyme ID | MGYG000001514_04050 | |||||||||||
| CAZy Family | GH11 | |||||||||||
| CAZyme Description | Bifunctional xylanase/deacetylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2810634; End: 2811701 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH11 | 38 | 219 | 2.4e-76 | 0.9887005649717514 |
| CBM36 | 241 | 354 | 1.4e-53 | 0.991304347826087 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00457 | Glyco_hydro_11 | 1.19e-88 | 38 | 218 | 1 | 175 | Glycosyl hydrolases family 11. |
| cd04078 | CBM36_xylanase-like | 5.62e-64 | 239 | 354 | 3 | 118 | Carbohydrate Binding Module family 36 (CBM36); appended mainly to glycoside hydrolase family 11 (GH11) domains; xylan binding. This family includes carbohydrate binding module family 36 (CBM36) most of which appear appended to glycoside hydrolase family 11 (GH11) domains. These CBMs are non-catalytic carbohydrate binding domains that facilitate the strong binding of the GH11 catalytic modules with their dedicated, insoluble substrates. GH11 domains have xylanase (endo-1,4-beta-xylanase) activity which catalyzes the hydrolysis of beta-1,4 bonds of xylan, the major component of hemicelluloses, to generate xylooligosaccharides and xylose. This family includes XynB from Dictyoglomus thermophilum Rt46B.1 and Xyn11A from Pseudobutyrivibrio xylanivorans Mz5T. Xyn11A is a multicatalytic enzyme with an N-terminal GH11 domain, a CBM36 domain, and a C-terminal putative NodB-like polysaccharide deacetylase which is predicted to be an acetyl esterase involved in debranching activity in the xylan backbone. CBM6 is an unusual CBM as it represents a chimera of two distinct binding sites with different modes of binding: binding site I within the loop regions and binding site II on the concave face of the beta-sandwich fold. Consistent with its structural and sequence similarity to CBM6, CBM36 binds xylan, but only at binding site I, and in a calcium-dependent manner; the latter suggests its potential application in affinity labeling. |
| pfam03422 | CBM_6 | 8.24e-07 | 241 | 354 | 1 | 123 | Carbohydrate binding module (family 6). |
| cd02795 | CBM6-CBM35-CBM36_like | 3.04e-05 | 239 | 354 | 1 | 124 | Carbohydrate Binding Module 6 (CBM6) and CBM35_like superfamily. Carbohydrate binding module family 6 (CBM6, family 6 CBM), also known as cellulose binding domain family VI (CBD VI), and related CBMs (CBM35 and CBM36). These are non-catalytic carbohydrate binding domains found in a range of enzymes that display activities against a diverse range of carbohydrate targets, including mannan, xylan, beta-glucans, cellulose, agarose, and arabinans. These domains facilitate the strong binding of the appended catalytic modules to their dedicated, insoluble substrates. Many of these CBMs are associated with glycoside hydrolase (GH) domains. CBM6 is an unusual CBM as it represents a chimera of two distinct binding sites with different modes of binding: binding site I within the loop regions and binding site II on the concave face of the beta-sandwich fold. CBM36s are calcium-dependent xylan binding domains. CBM35s display conserved specificity through extensive sequence similarity, but divergent function through their appended catalytic modules. This alignment model also contains the C-terminal domains of bacterial insecticidal toxins, where they may be involved in determining insect specificity through carbohydrate binding functionality. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AZK47932.1 | 8.28e-250 | 1 | 355 | 1 | 354 |
| AVI01407.1 | 5.77e-246 | 1 | 355 | 1 | 356 |
| QXF28299.1 | 3.54e-213 | 4 | 354 | 16 | 376 |
| ABB77852.1 | 3.38e-211 | 4 | 354 | 16 | 376 |
| CAM82403.1 | 1.30e-210 | 4 | 354 | 5 | 365 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6KJL_A | 1.34e-195 | 27 | 354 | 1 | 327 | XylanaseJ from Bacillus sp. strain 41M-1 [Bacillus sp. 41M-1],6KJL_B Xylanase J from Bacillus sp. strain 41M-1 [Bacillus sp. 41M-1] |
| 2DCJ_A | 4.92e-195 | 29 | 354 | 29 | 353 | Atwo-domain structure of alkaliphilic XynJ from Bacillus sp. 41M-1 [Bacillus sp. 41M-1],2DCJ_B A two-domain structure of alkaliphilic XynJ from Bacillus sp. 41M-1 [Bacillus sp. 41M-1],2DCK_A A tetragonal-lattice structure of alkaliphilic XynJ from Bacillus sp. 41M-1 [Bacillus sp. 41M-1] |
| 6KKA_A | 1.06e-194 | 29 | 354 | 2 | 326 | XylanaseJ mutant from Bacillus sp. 41M-1 [Bacillus sp. 41M-1],6KKA_B Xylanase J mutant from Bacillus sp. 41M-1 [Bacillus sp. 41M-1] |
| 7AYP_A | 8.36e-171 | 26 | 354 | 1 | 352 | Structureof a GH11 domain refined from the X-ray diffraction data of a GH11-CBM36-1 crystal. [uncultured bacterium] |
| 2F6B_A | 3.04e-125 | 29 | 228 | 3 | 203 | Structuraland active site modification studies implicate Glu, Trp and Arg in the activity of xylanase from alkalophilic Bacillus sp. (NCL 87-6-10). [Bacillus],2F6B_B Structural and active site modification studies implicate Glu, Trp and Arg in the activity of xylanase from alkalophilic Bacillus sp. (NCL 87-6-10). [Bacillus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P83513 | 2.74e-166 | 26 | 351 | 15 | 362 | Bifunctional xylanase/deacetylase OS=Pseudobutyrivibrio xylanivorans OX=185007 GN=xyn11A PE=1 SV=2 |
| Q8GJ44 | 9.40e-109 | 4 | 342 | 3 | 366 | Endo-1,4-beta-xylanase A OS=Thermoclostridium stercorarium OX=1510 GN=xynA PE=1 SV=2 |
| P33558 | 1.79e-105 | 4 | 342 | 3 | 367 | Endo-1,4-beta-xylanase A OS=Thermoclostridium stercorarium OX=1510 GN=xynA PE=1 SV=2 |
| P17137 | 3.97e-102 | 27 | 227 | 60 | 260 | Endo-1,4-beta-xylanase OS=Clostridium saccharobutylicum OX=169679 GN=xynB PE=3 SV=1 |
| P00694 | 7.36e-102 | 1 | 227 | 1 | 227 | Endo-1,4-beta-xylanase A OS=Bacillus pumilus OX=1408 GN=xynA PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
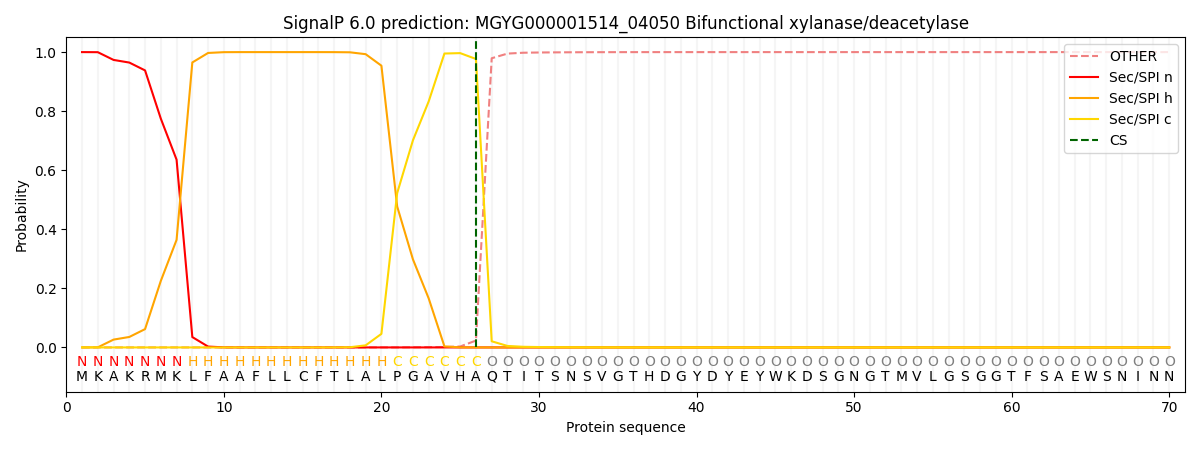
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000238 | 0.999051 | 0.000184 | 0.000182 | 0.000164 | 0.000151 |