You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001514_04369
You are here: Home > Sequence: MGYG000001514_04369
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
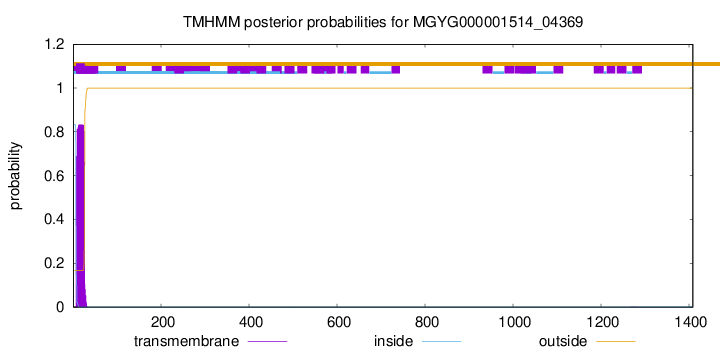
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A ihumii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A ihumii | |||||||||||
| CAZyme ID | MGYG000001514_04369 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3153773; End: 3158002 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM51 | 43 | 188 | 2.9e-36 | 0.9850746268656716 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14244 | GH_101_like | 8.90e-60 | 472 | 779 | 2 | 298 | Endo-a-N-acetylgalactosaminidase and related glcyosyl hydrolases. This family contains the enzymatically active domain of cell surface proteins that specifically cleave Gal-beta-1,3-GalNAc-alpha-Ser/Thr (T-antigen, galacto-N-biose), the core 1 type O-linked glycan common to mucin glycoproteins (EC:3.2.1.97). It has been classified as glycosyl hydrolase family 101 in the Cazy resource. Virulence of pathogenic organisms such as the Gram-positive Streptococcus pneumoniae and other commensal human bacteria is largely determined by their ability to degrade host glycoproteins and to metabolize the resultant carbohydrates. |
| pfam08305 | NPCBM | 3.48e-41 | 40 | 189 | 1 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 3.14e-30 | 41 | 189 | 4 | 145 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| NF033190 | inl_like_NEAT_1 | 3.67e-13 | 1112 | 1384 | 446 | 733 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| pfam00395 | SLH | 3.46e-07 | 1286 | 1327 | 1 | 42 | S-layer homology domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AZS16787.1 | 0.0 | 1 | 696 | 1 | 696 |
| QTH41000.1 | 1.49e-153 | 357 | 1405 | 243 | 1445 |
| SYX82446.1 | 5.14e-133 | 361 | 894 | 32 | 569 |
| SMF83630.1 | 3.26e-130 | 361 | 894 | 32 | 569 |
| QJD87425.1 | 1.27e-129 | 43 | 894 | 261 | 1043 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6BT4_A | 1.43e-15 | 1227 | 1409 | 26 | 202 | Crystalstructure of the SLH domain of Sap from Bacillus anthracis in complex with a pyruvylated SCWP unit [Bacillus anthracis] |
| 3PYW_A | 1.47e-15 | 1227 | 1409 | 5 | 181 | Thestructure of the SLH domain from B. anthracis surface array protein at 1.8A [Bacillus anthracis] |
| 2VMG_A | 2.39e-11 | 32 | 174 | 5 | 147 | Thestructure of CBM51 from Clostridium perfringens GH95 in complex with methyl-galactose [Clostridium perfringens] |
| 2VMH_A | 7.12e-11 | 43 | 174 | 10 | 141 | Thestructure of CBM51 from Clostridium perfringens GH95 [Clostridium perfringens] |
| 2VMI_A | 9.69e-11 | 43 | 174 | 10 | 141 | Thestructure of seleno-methionine labelled CBM51 from Clostridium perfringens GH95 [Clostridium perfringens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| C6CRV0 | 1.48e-28 | 909 | 1405 | 966 | 1461 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
| P38536 | 3.26e-23 | 1170 | 1402 | 1624 | 1854 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| P38535 | 5.51e-23 | 1219 | 1402 | 900 | 1080 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
| P38537 | 3.96e-20 | 1235 | 1401 | 42 | 204 | Surface-layer 125 kDa protein OS=Lysinibacillus sphaericus OX=1421 PE=3 SV=1 |
| P19424 | 2.33e-17 | 1227 | 1402 | 41 | 213 | Endoglucanase OS=Bacillus sp. (strain KSM-635) OX=1415 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
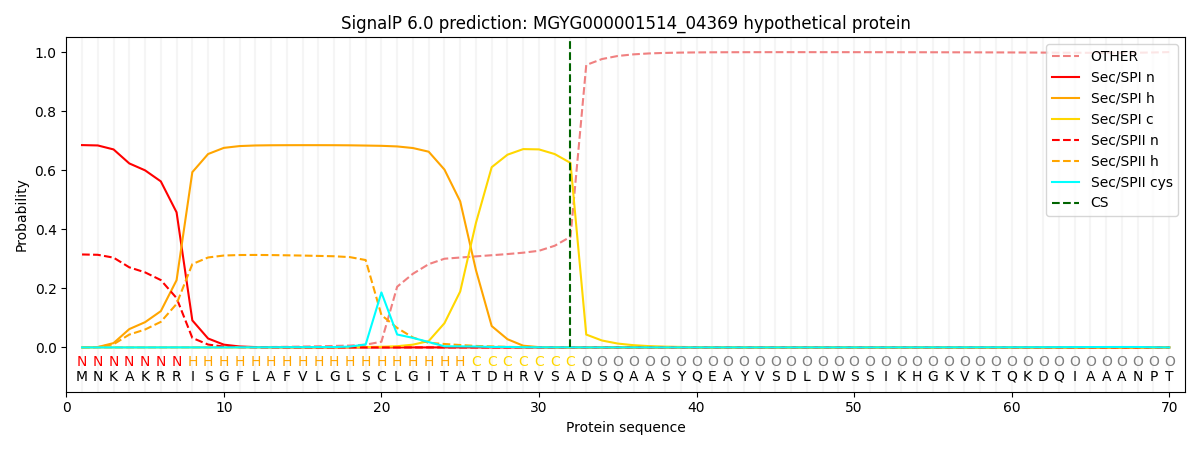
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000746 | 0.673540 | 0.324671 | 0.000376 | 0.000361 | 0.000269 |