You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001514_04992
You are here: Home > Sequence: MGYG000001514_04992
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A ihumii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A ihumii | |||||||||||
| CAZyme ID | MGYG000001514_04992 | |||||||||||
| CAZy Family | CBM66 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3801961; End: 3803841 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 241 | 613 | 7.7e-139 | 0.9893333333333333 |
| CBM66 | 54 | 211 | 5.8e-23 | 0.9870967741935484 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13229 | Beta_helix | 4.70e-08 | 329 | 521 | 2 | 157 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13385 | Laminin_G_3 | 1.25e-04 | 102 | 191 | 25 | 109 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| smart00282 | LamG | 7.26e-04 | 120 | 188 | 21 | 86 | Laminin G domain. |
| pfam02210 | Laminin_G_2 | 0.005 | 114 | 181 | 12 | 73 | Laminin G domain. This family includes the Thrombospondin N-terminal-like domain, a Laminin G subfamily. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QKS46472.1 | 3.30e-287 | 37 | 626 | 4 | 590 |
| CQR55664.1 | 6.09e-285 | 32 | 625 | 24 | 645 |
| QYR21238.1 | 4.66e-250 | 42 | 625 | 9 | 603 |
| APO48380.1 | 2.50e-239 | 240 | 626 | 16 | 402 |
| QZN75200.1 | 2.51e-239 | 240 | 626 | 73 | 459 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1RU4_A | 4.08e-54 | 245 | 616 | 18 | 379 | ChainA, Pectate lyase [Dickeya chrysanthemi] |
| 5OLQ_A | 1.47e-17 | 245 | 528 | 6 | 322 | Rhamnogalacturonanlyase [Bacteroides thetaiotaomicron],5OLQ_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLQ_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLS_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron] |
| 4AZZ_A | 6.58e-06 | 54 | 216 | 8 | 165 | Carbohydratebinding module CBM66 from Bacillus subtilis [Bacillus subtilis],4AZZ_B Carbohydrate binding module CBM66 from Bacillus subtilis [Bacillus subtilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0C1A7 | 4.06e-53 | 245 | 616 | 43 | 404 | Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
| P0C1A6 | 4.01e-52 | 245 | 616 | 43 | 404 | Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
| P22751 | 1.08e-43 | 243 | 546 | 390 | 667 | Pectate disaccharide-lyase OS=Dickeya chrysanthemi OX=556 GN=pelX PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
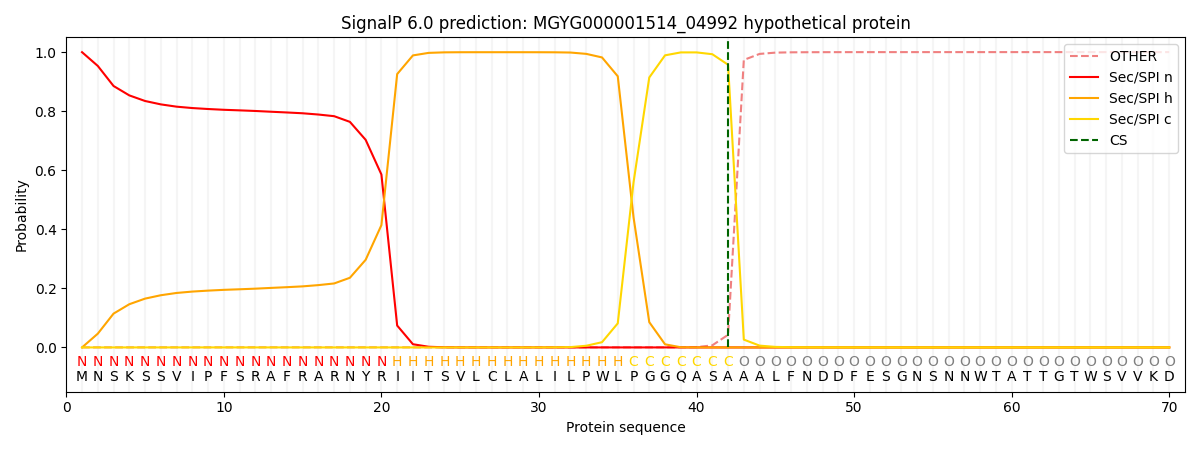
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000759 | 0.998425 | 0.000246 | 0.000204 | 0.000165 | 0.000156 |

