You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001518_00803
You are here: Home > Sequence: MGYG000001518_00803
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
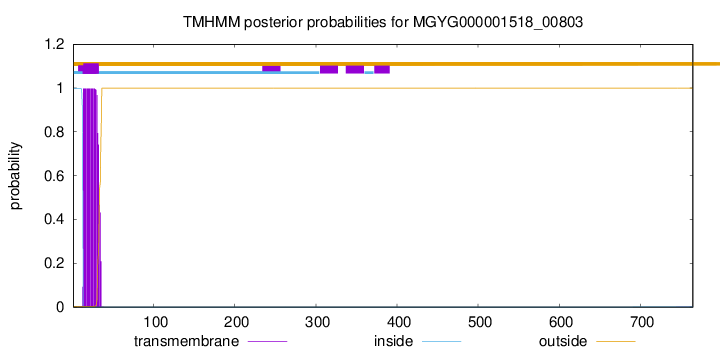
TMHMM annotations
Basic Information help
| Species | Gorillibacterium timonense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Gorillibacterium; Gorillibacterium timonense | |||||||||||
| CAZyme ID | MGYG000001518_00803 | |||||||||||
| CAZy Family | CBM22 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 440802; End: 443099 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 200 | 522 | 2.5e-117 | 0.9933993399339934 |
| CBM22 | 39 | 168 | 8.1e-33 | 0.9770992366412213 |
| CBM3 | 624 | 703 | 2.9e-23 | 0.9886363636363636 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 8.49e-133 | 200 | 521 | 1 | 310 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 2.21e-129 | 241 | 519 | 1 | 263 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 9.64e-103 | 226 | 527 | 52 | 345 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam02018 | CBM_4_9 | 4.82e-23 | 38 | 170 | 3 | 133 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
| pfam00942 | CBM_3 | 5.38e-23 | 623 | 702 | 1 | 82 | Cellulose binding domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWV32895.1 | 0.0 | 11 | 765 | 7 | 770 |
| ANY65885.1 | 0.0 | 11 | 765 | 10 | 781 |
| APO45385.1 | 1.15e-309 | 11 | 765 | 7 | 775 |
| AEI43166.1 | 2.80e-291 | 20 | 765 | 16 | 672 |
| AAF61649.1 | 6.66e-287 | 12 | 765 | 11 | 712 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6D5C_A | 1.44e-111 | 196 | 522 | 19 | 350 | Structureof Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5C_B Structure of Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5C_C Structure of Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii] |
| 5OFJ_A | 1.20e-109 | 197 | 523 | 8 | 339 | Crystalstructure of N-terminal domain of bifunctional CbXyn10C [Caldicellulosiruptor bescii DSM 6725] |
| 5OFK_A | 9.32e-109 | 197 | 523 | 8 | 339 | Crystalstructure of CbXyn10C variant E140Q/E248Q complexed with xyloheptaose [Caldicellulosiruptor bescii DSM 6725],5OFL_A Crystal structure of CbXyn10C variant E140Q/E248Q complexed with cellohexaose [Caldicellulosiruptor bescii DSM 6725] |
| 7NL2_A | 2.40e-102 | 189 | 525 | 1 | 343 | ChainA, Beta-xylanase [Pseudothermotoga thermarum DSM 5069],7NL2_B Chain B, Beta-xylanase [Pseudothermotoga thermarum DSM 5069] |
| 2Q8X_A | 1.67e-98 | 196 | 520 | 4 | 329 | Thehigh-resolution crystal structure of ixt6, a thermophilic, intracellular xylanase from G. stearothermophilus [unidentified],2Q8X_B The high-resolution crystal structure of ixt6, a thermophilic, intracellular xylanase from G. stearothermophilus [unidentified],3MSD_A Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MSD_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MSG_A Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MSG_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P10474 | 1.40e-133 | 196 | 765 | 41 | 570 | Endoglucanase/exoglucanase B OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=celB PE=3 SV=1 |
| Q60042 | 4.05e-113 | 29 | 520 | 191 | 686 | Endo-1,4-beta-xylanase A OS=Thermotoga neapolitana OX=2337 GN=xynA PE=1 SV=1 |
| Q60037 | 1.69e-110 | 29 | 520 | 196 | 690 | Endo-1,4-beta-xylanase A OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=xynA PE=1 SV=1 |
| P36917 | 1.10e-102 | 30 | 527 | 190 | 680 | Endo-1,4-beta-xylanase A OS=Thermoanaerobacterium saccharolyticum OX=28896 GN=xynA PE=1 SV=1 |
| P38535 | 9.27e-98 | 25 | 654 | 26 | 636 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
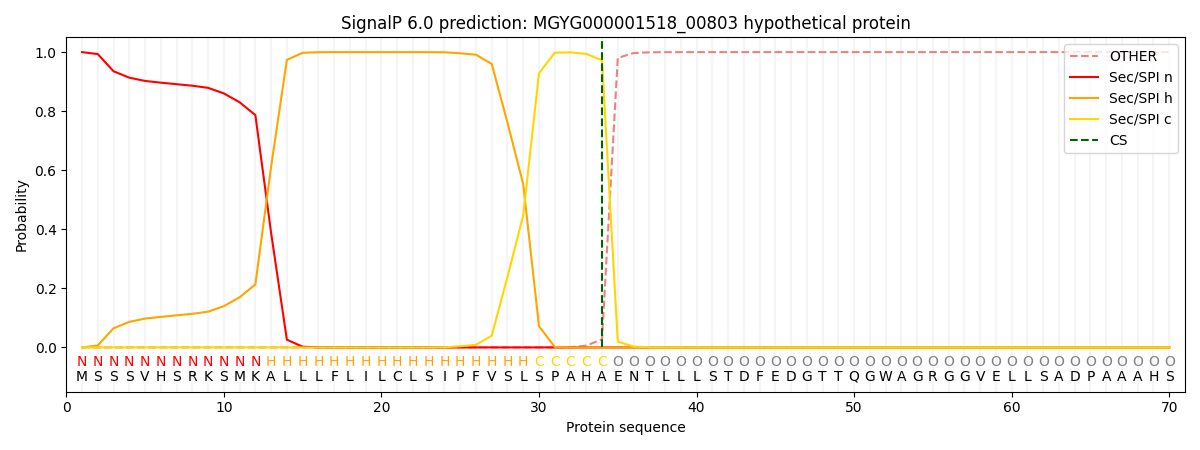
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000272 | 0.999039 | 0.000211 | 0.000165 | 0.000151 | 0.000135 |