You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001518_02801
You are here: Home > Sequence: MGYG000001518_02801
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Gorillibacterium timonense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Gorillibacterium; Gorillibacterium timonense | |||||||||||
| CAZyme ID | MGYG000001518_02801 | |||||||||||
| CAZy Family | SLH | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 243952; End: 248871 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| SLH | 1463 | 1503 | 6.9e-16 | 0.9761904761904762 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033190 | inl_like_NEAT_1 | 7.99e-20 | 1439 | 1634 | 559 | 752 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| NF033190 | inl_like_NEAT_1 | 1.05e-13 | 1406 | 1573 | 565 | 753 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| pfam00395 | SLH | 6.90e-11 | 1463 | 1503 | 1 | 42 | S-layer homology domain. |
| pfam00395 | SLH | 1.57e-10 | 1522 | 1564 | 1 | 42 | S-layer homology domain. |
| smart00060 | FN3 | 2.15e-10 | 1201 | 1281 | 1 | 83 | Fibronectin type 3 domain. One of three types of internal repeat within the plasma protein, fibronectin. The tenth fibronectin type III repeat contains a RGD cell recognition sequence in a flexible loop between 2 strands. Type III modules are present in both extracellular and intracellular proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QMV41393.1 | 7.39e-185 | 1001 | 1635 | 1598 | 2235 |
| QNK59241.1 | 1.64e-69 | 1311 | 1635 | 1134 | 1459 |
| ASR48635.1 | 1.73e-67 | 1005 | 1634 | 157 | 765 |
| AET58457.1 | 1.34e-63 | 1005 | 1634 | 159 | 770 |
| BBI31951.1 | 1.29e-62 | 1309 | 1633 | 1212 | 1530 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6BT4_A | 6.76e-16 | 1462 | 1632 | 25 | 194 | Crystalstructure of the SLH domain of Sap from Bacillus anthracis in complex with a pyruvylated SCWP unit [Bacillus anthracis] |
| 3PYW_A | 6.91e-16 | 1462 | 1632 | 4 | 173 | Thestructure of the SLH domain from B. anthracis surface array protein at 1.8A [Bacillus anthracis] |
| 4AQ1_A | 8.07e-07 | 1462 | 1634 | 3 | 168 | Structureof the SbsB S-layer protein of Geobacillus stearothermophilus PV72p2 in complex with nanobody KB6 [Geobacillus stearothermophilus] |
| 4AQ1_C | 8.07e-07 | 1462 | 1634 | 3 | 168 | Structureof the SbsB S-layer protein of Geobacillus stearothermophilus PV72p2 in complex with nanobody KB6 [Geobacillus stearothermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P38536 | 3.73e-30 | 1389 | 1637 | 1620 | 1858 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| P38535 | 3.92e-25 | 1460 | 1638 | 905 | 1085 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
| C6CRV0 | 3.19e-20 | 1390 | 1632 | 1218 | 1457 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
| P19424 | 4.82e-19 | 1455 | 1630 | 33 | 210 | Endoglucanase OS=Bacillus sp. (strain KSM-635) OX=1415 PE=1 SV=1 |
| Q06853 | 3.62e-17 | 1461 | 1572 | 523 | 638 | Cell surface glycoprotein 2 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=Cthe_3079 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
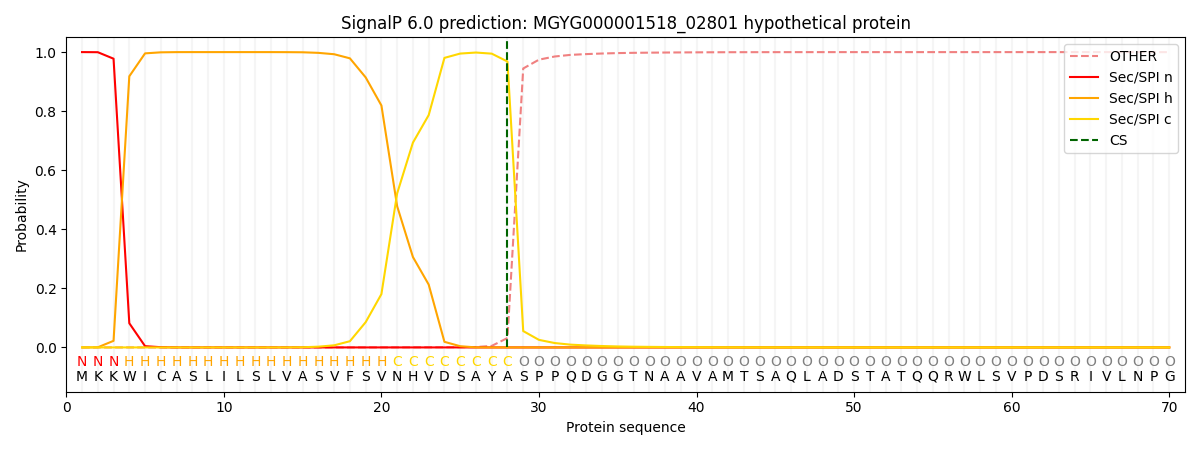
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000305 | 0.998941 | 0.000207 | 0.000197 | 0.000159 | 0.000146 |
