You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001518_03293
You are here: Home > Sequence: MGYG000001518_03293
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Gorillibacterium timonense | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Gorillibacterium; Gorillibacterium timonense | |||||||||||
| CAZyme ID | MGYG000001518_03293 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 838843; End: 840594 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 46 | 359 | 1.7e-135 | 0.9967741935483871 |
| CBM3 | 447 | 518 | 4.3e-18 | 0.9772727272727273 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG2730 | BglC | 4.97e-55 | 41 | 381 | 34 | 392 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam00150 | Cellulase | 2.84e-49 | 45 | 354 | 1 | 272 | Cellulase (glycosyl hydrolase family 5). |
| pfam00942 | CBM_3 | 2.40e-19 | 446 | 518 | 1 | 82 | Cellulose binding domain. |
| smart01067 | CBM_3 | 3.50e-17 | 446 | 519 | 1 | 83 | Cellulose binding domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AZS14931.1 | 7.99e-284 | 1 | 583 | 1 | 629 |
| QJD87157.1 | 4.56e-267 | 9 | 583 | 17 | 632 |
| ASA23029.1 | 3.44e-248 | 3 | 583 | 2 | 557 |
| QKS44641.1 | 1.44e-245 | 3 | 583 | 7 | 729 |
| AIQ27461.1 | 4.76e-245 | 2 | 583 | 5 | 554 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1VRX_A | 7.61e-135 | 32 | 376 | 1 | 347 | ChainA, ENDOCELLULASE E1 FROM A. CELLULOLYTICUS [Acidothermus cellulolyticus],1VRX_B Chain B, ENDOCELLULASE E1 FROM A. CELLULOLYTICUS [Acidothermus cellulolyticus] |
| 1ECE_A | 6.11e-134 | 32 | 376 | 1 | 347 | AcidothermusCellulolyticus Endocellulase E1 Catalytic Domain In Complex With A Cellotetraose [Acidothermus cellulolyticus],1ECE_B Acidothermus Cellulolyticus Endocellulase E1 Catalytic Domain In Complex With A Cellotetraose [Acidothermus cellulolyticus] |
| 4TUF_A | 4.65e-104 | 43 | 379 | 8 | 343 | Catalyticdomain of the major endoglucanase from Xanthomonas campestris pv. campestris [Xanthomonas campestris pv. campestris str. ATCC 33913],4TUF_B Catalytic domain of the major endoglucanase from Xanthomonas campestris pv. campestris [Xanthomonas campestris pv. campestris str. ATCC 33913],4TUF_C Catalytic domain of the major endoglucanase from Xanthomonas campestris pv. campestris [Xanthomonas campestris pv. campestris str. ATCC 33913],4TUF_D Catalytic domain of the major endoglucanase from Xanthomonas campestris pv. campestris [Xanthomonas campestris pv. campestris str. ATCC 33913] |
| 3W6M_A | 9.01e-94 | 43 | 380 | 19 | 376 | Contributionof disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6M_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6M_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM2_A Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM2_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM2_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3] |
| 3VVG_A | 9.01e-94 | 43 | 380 | 19 | 376 | TheCrystal Structure of Cellulase-Inhibitor Complex. [Pyrococcus horikoshii OT3],3VVG_B The Crystal Structure of Cellulase-Inhibitor Complex. [Pyrococcus horikoshii OT3],3VVG_C The Crystal Structure of Cellulase-Inhibitor Complex. [Pyrococcus horikoshii OT3],3W6L_A Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6L_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],3W6L_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM1_A Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM1_B Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3],4DM1_C Contribution of disulfide bond toward thermostability in hyperthermostable endocellulase [Pyrococcus horikoshii OT3] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23548 | 2.28e-202 | 8 | 383 | 10 | 385 | Endoglucanase OS=Paenibacillus polymyxa OX=1406 PE=3 SV=2 |
| P54583 | 1.81e-130 | 31 | 376 | 41 | 388 | Endoglucanase E1 OS=Acidothermus cellulolyticus (strain ATCC 43068 / DSM 8971 / 11B) OX=351607 GN=Acel_0614 PE=1 SV=1 |
| P19487 | 2.41e-102 | 20 | 379 | 6 | 368 | Major extracellular endoglucanase OS=Xanthomonas campestris pv. campestris (strain ATCC 33913 / DSM 3586 / NCPPB 528 / LMG 568 / P 25) OX=190485 GN=engXCA PE=1 SV=2 |
| P50400 | 2.99e-78 | 7 | 376 | 14 | 409 | Endoglucanase D OS=Cellulomonas fimi OX=1708 GN=cenD PE=3 SV=1 |
| Q05332 | 8.01e-74 | 3 | 376 | 5 | 440 | Endoglucanase G OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celG PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
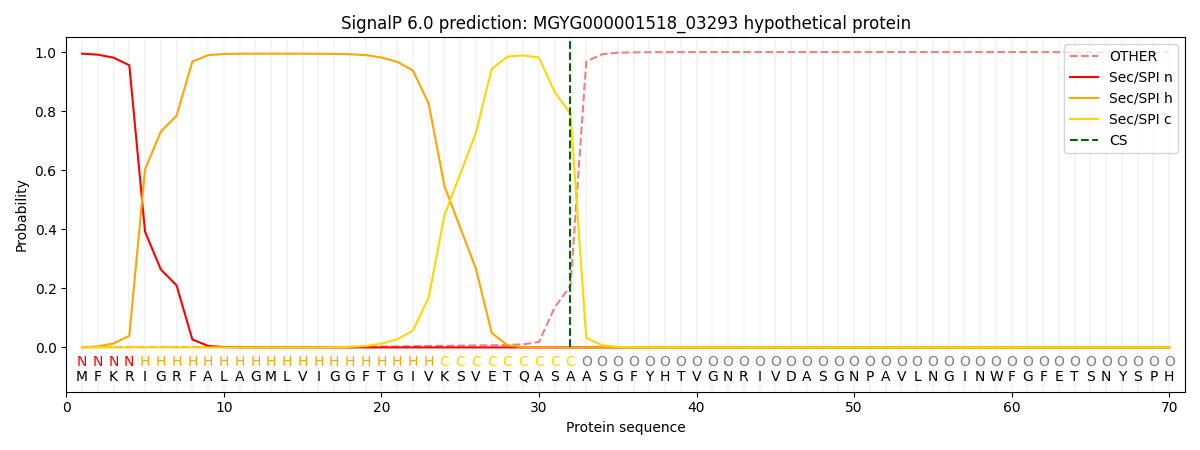
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.004696 | 0.989516 | 0.004753 | 0.000468 | 0.000299 | 0.000274 |
