You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001522_01302
You are here: Home > Sequence: MGYG000001522_01302
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pauljensenia radingae_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; Pauljensenia; Pauljensenia radingae_A | |||||||||||
| CAZyme ID | MGYG000001522_01302 | |||||||||||
| CAZy Family | CE5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 383845; End: 384798 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE5 | 78 | 282 | 1.3e-36 | 0.9947089947089947 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01083 | Cutinase | 2.78e-14 | 119 | 282 | 43 | 173 | Cutinase. |
| pfam08237 | PE-PPE | 0.001 | 114 | 168 | 2 | 63 | PE-PPE domain. This domain is found C terminal to the PE (pfam00934) and PPE (pfam00823) domains. The secondary structure of this domain is predicted to be a mixture of alpha helices and beta strands. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SDU01509.1 | 1.57e-210 | 1 | 317 | 1 | 317 |
| QZY52027.1 | 4.38e-47 | 8 | 282 | 6 | 260 |
| QIM16497.1 | 1.74e-41 | 8 | 288 | 18 | 289 |
| QIK61901.1 | 5.82e-41 | 1 | 282 | 1 | 274 |
| ACY97091.1 | 6.53e-33 | 79 | 282 | 35 | 235 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1BS9_A | 1.81e-08 | 79 | 270 | 2 | 199 | AcetylxylanEsterase From P. Purpurogenum Refined At 1.10 Angstroms [Talaromyces purpureogenus] |
| 1G66_A | 1.81e-08 | 79 | 270 | 2 | 199 | ACETYLXYLANESTERASE AT 0.90 ANGSTROM RESOLUTION [Talaromyces purpureogenus] |
| 2AXE_A | 4.49e-08 | 79 | 270 | 2 | 199 | IodinatedComplex Of Acetyl Xylan Esterase At 1.80 Angstroms [Talaromyces purpureogenus] |
| 1QOZ_A | 6.08e-08 | 79 | 265 | 2 | 190 | ChainA, ACETYL XYLAN ESTERASE [Trichoderma reesei],1QOZ_B Chain B, ACETYL XYLAN ESTERASE [Trichoderma reesei] |
| 1CUX_A | 2.20e-07 | 79 | 276 | 31 | 199 | ChainA, CUTINASE [Fusarium vanettenii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P63880 | 7.56e-14 | 66 | 282 | 22 | 213 | Carboxylesterase Culp1 OS=Mycobacterium bovis (strain ATCC BAA-935 / AF2122/97) OX=233413 GN=BQ2027_MB2006C PE=3 SV=1 |
| P9WP42 | 7.56e-14 | 66 | 282 | 22 | 213 | Carboxylesterase Culp1 OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=MT2037 PE=3 SV=1 |
| P9WP43 | 7.56e-14 | 66 | 282 | 22 | 213 | Carboxylesterase Culp1 OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=cut7 PE=1 SV=1 |
| A6WFI5 | 6.03e-12 | 79 | 278 | 36 | 214 | Cutinase OS=Kineococcus radiotolerans (strain ATCC BAA-149 / DSM 14245 / SRS30216) OX=266940 GN=cut PE=1 SV=1 |
| O06319 | 3.68e-08 | 79 | 282 | 47 | 226 | Phospholipase Culp4 OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=cut4 PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
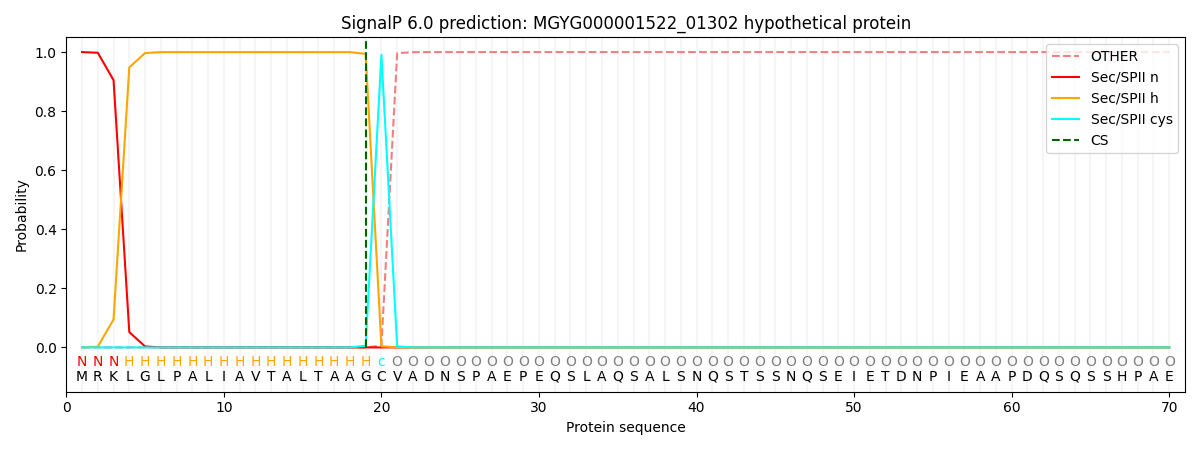
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000062 | 0.000000 | 0.000000 | 0.000000 |
