You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001522_01397
You are here: Home > Sequence: MGYG000001522_01397
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
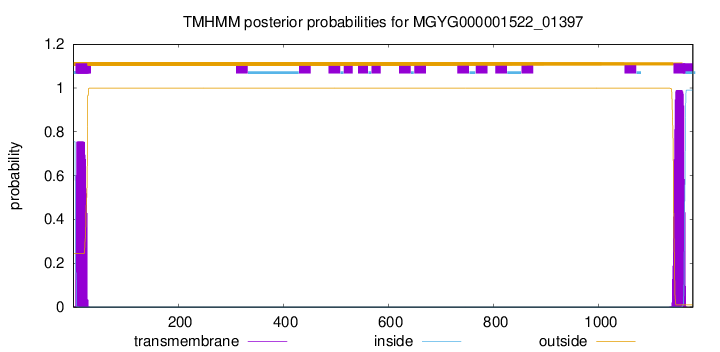
TMHMM annotations
Basic Information help
| Species | Pauljensenia radingae_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; Pauljensenia; Pauljensenia radingae_A | |||||||||||
| CAZyme ID | MGYG000001522_01397 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 494723; End: 498262 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 42 | 554 | 1e-174 | 0.9978070175438597 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14587 | Glyco_hydr_30_2 | 5.14e-18 | 42 | 405 | 6 | 364 | O-Glycosyl hydrolase family 30. |
| pfam14200 | RicinB_lectin_2 | 8.43e-06 | 652 | 709 | 5 | 62 | Ricin-type beta-trefoil lectin domain-like. |
| NF035930 | lectin_2 | 6.37e-04 | 662 | 699 | 155 | 196 | lectin. Lectins are important adhesin proteins, which bind carbohydrate structures on host cell surface. The carbohydrate specificity of diverse lectins to a large extent dictates bacteria tissue tropism by mediating specific attachment to unique host sites expressing the corresponding carbohydrate receptor. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CED92153.1 | 0.0 | 39 | 1067 | 35 | 1062 |
| QIG38692.1 | 0.0 | 42 | 1099 | 38 | 1098 |
| QCQ17871.1 | 0.0 | 8 | 1091 | 6 | 1093 |
| BAJ75662.1 | 0.0 | 6 | 1087 | 5 | 1088 |
| AXL13272.1 | 0.0 | 1 | 1081 | 1 | 1072 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A0A401ETL2 | 2.22e-120 | 38 | 936 | 35 | 1045 | Exo-beta-1,6-galactobiohydrolase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=bl1,6Gal PE=1 SV=1 |
| Q76FP5 | 5.09e-35 | 22 | 487 | 6 | 409 | Endo-beta-1,6-galactanase OS=Hypocrea rufa OX=5547 GN=6GAL PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
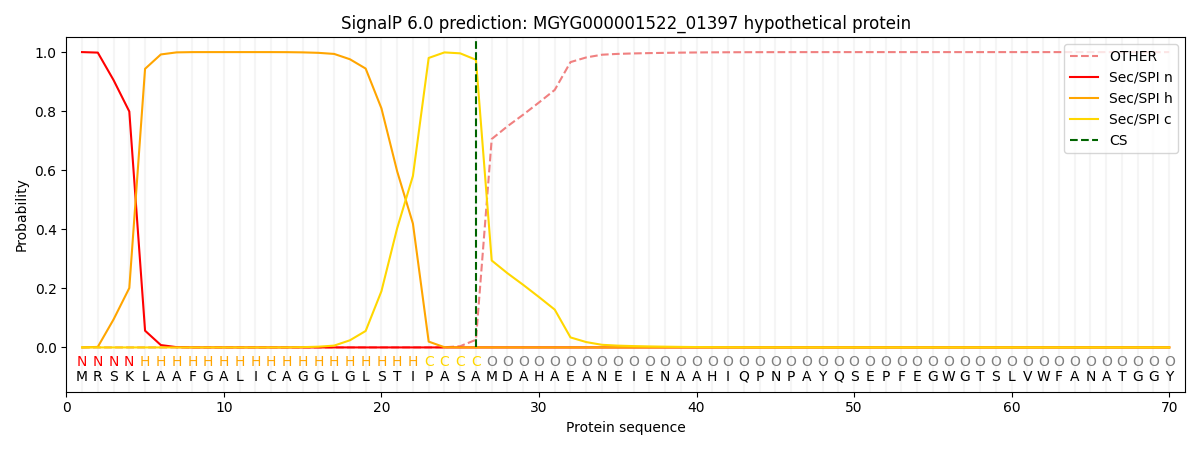
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000377 | 0.998794 | 0.000202 | 0.000259 | 0.000192 | 0.000175 |