You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001522_01796
You are here: Home > Sequence: MGYG000001522_01796
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pauljensenia radingae_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Actinomycetaceae; Pauljensenia; Pauljensenia radingae_A | |||||||||||
| CAZyme ID | MGYG000001522_01796 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 946590; End: 947786 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 21 | 302 | 4.3e-119 | 0.9964539007092199 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3934 | COG3934 | 0.001 | 8 | 335 | 5 | 318 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
| cd19608 | GH113_mannanase-like | 0.003 | 10 | 158 | 5 | 145 | Glycoside hydrolase family 113 beta-1,4-mannanase and similar proteins. Mannan endo-1,4-beta mannosidase (E.C 3.2.1.78) randomly cleaves (1->4)-beta-D-mannosidic linkages in mannans, galactomannans and glucomannans and is also called beta-1,4-mannanase, endo-1,4-beta-mannanase, endo-beta-1,4-mannase, beta-mannanase B, beta-1, 4-mannan 4-mannanohydrolase, endo-beta-mannanase, beta-D-mannanase, 1,4-beta-D-mannan mannanohydrolase, and 4-beta-D-mannan mannanohydrolase. (1->4)-beta-linked mannans are polysaccharides with a linear polymer backbone of (1->4)-beta-linked mannose units (in plants and fungi) or alternating mannose and glucose/galactose units (glucomannan in plants and fungi, and galactomannan and galactoglucomannan in plants), such as in the hemicellulose fraction of hard- and softwoods. Complete degradation of mannan requires a series of enzymes, including beta-1,4-mannanase. According to the CAZy database beta-1,4-mannanases are grouped into various glycoside hydrolase (GH) families; GH family 113 beta-1,4-mannanases include mostly bacterial and archaeal sequences. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SDU07880.1 | 4.53e-310 | 1 | 398 | 1 | 398 |
| SDU78319.1 | 1.88e-146 | 9 | 398 | 3 | 398 |
| QJC21085.1 | 1.01e-144 | 9 | 378 | 3 | 375 |
| ADH91800.1 | 1.10e-143 | 9 | 395 | 3 | 411 |
| SQH27378.1 | 1.10e-143 | 9 | 395 | 3 | 411 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7LR7_A | 2.24e-129 | 3 | 398 | 10 | 404 | ChainA, Uncharacterized protein ScGH5_18 [Streptomyces cattleya NRRL 8057 = DSM 46488] |
| 7LR8_A | 1.81e-128 | 3 | 398 | 10 | 404 | ChainA, Uncharacterized protein ScGH5_18 [Streptomyces cattleya NRRL 8057 = DSM 46488],7LR8_B Chain B, Uncharacterized protein ScGH5_18 [Streptomyces cattleya NRRL 8057 = DSM 46488] |
| 6GVB_A | 1.25e-124 | 10 | 335 | 4 | 333 | ChainA, exo-beta-1,4-mannosidase [Cutibacterium acnes],6GVB_B Chain B, exo-beta-1,4-mannosidase [Cutibacterium acnes],6GVB_C Chain C, exo-beta-1,4-mannosidase [Cutibacterium acnes],6GVB_D Chain D, exo-beta-1,4-mannosidase [Cutibacterium acnes] |
| 7LQX_A | 7.25e-111 | 2 | 383 | 19 | 396 | ChainA, Glycosyl hydrolase BlGH5_18 [Bifidobacterium longum subsp. infantis ATCC 15697 = JCM 1222 = DSM 20088] |
| 7LR1_A | 1.49e-100 | 6 | 383 | 20 | 393 | ChainA, Glycosyl hydrolase BlGH5_18 [Bifidobacterium longum subsp. longum ATCC 55813],7LR1_B Chain B, Glycosyl hydrolase BlGH5_18 [Bifidobacterium longum subsp. longum ATCC 55813],7LR1_C Chain C, Glycosyl hydrolase BlGH5_18 [Bifidobacterium longum subsp. longum ATCC 55813],7LR1_D Chain D, Glycosyl hydrolase BlGH5_18 [Bifidobacterium longum subsp. longum ATCC 55813],7LR2_A Chain A, Glycosyl hydrolase BlGH5_18 [Bifidobacterium longum subsp. longum ATCC 55813],7LR2_B Chain B, Glycosyl hydrolase BlGH5_18 [Bifidobacterium longum subsp. longum ATCC 55813],7LR2_C Chain C, Glycosyl hydrolase BlGH5_18 [Bifidobacterium longum subsp. longum ATCC 55813],7LR2_D Chain D, Glycosyl hydrolase BlGH5_18 [Bifidobacterium longum subsp. longum ATCC 55813] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
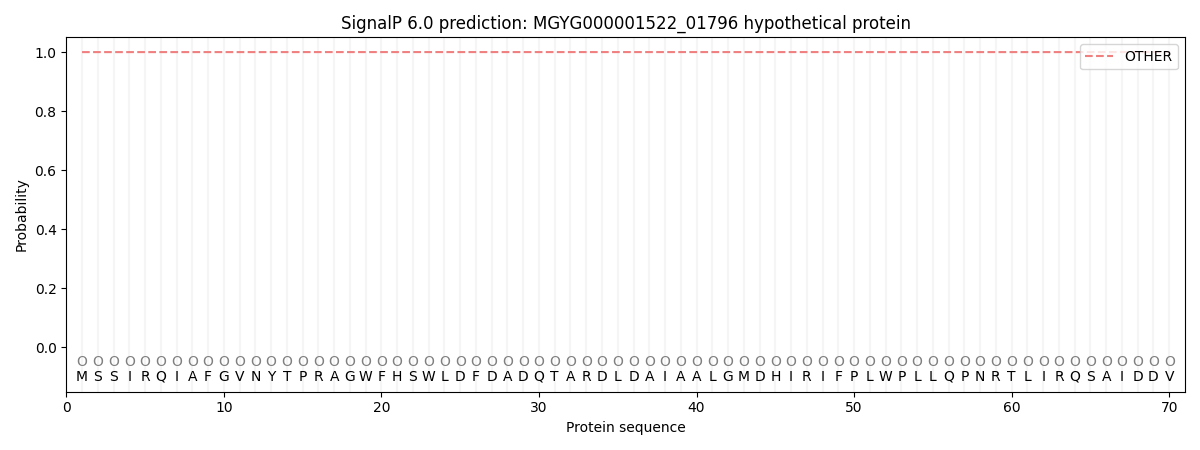
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000068 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
