You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001523_01077
You are here: Home > Sequence: MGYG000001523_01077
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
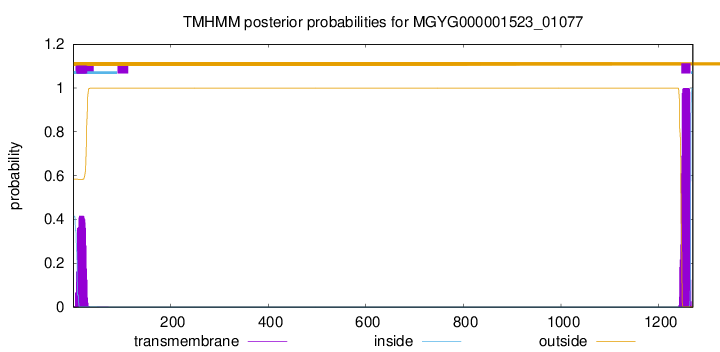
TMHMM annotations
Basic Information help
| Species | Gracilibacillus massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales_D; Amphibacillaceae; Gracilibacillus; Gracilibacillus massiliensis | |||||||||||
| CAZyme ID | MGYG000001523_01077 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 315606; End: 319418 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 44 | 515 | 1.1e-174 | 0.9978070175438597 |
| CBM13 | 533 | 679 | 9.8e-16 | 0.7712765957446809 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14587 | Glyco_hydr_30_2 | 3.42e-37 | 43 | 376 | 3 | 356 | O-Glycosyl hydrolase family 30. |
| pfam14200 | RicinB_lectin_2 | 1.17e-13 | 564 | 651 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| COG5520 | XynC | 2.68e-12 | 15 | 517 | 12 | 428 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| pfam07532 | Big_4 | 1.88e-11 | 672 | 729 | 2 | 59 | Bacterial Ig-like domain (group 4). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
| pfam14200 | RicinB_lectin_2 | 1.36e-09 | 610 | 663 | 1 | 54 | Ricin-type beta-trefoil lectin domain-like. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGQ44778.1 | 0.0 | 5 | 1047 | 19 | 1077 |
| QNF28725.1 | 0.0 | 24 | 1047 | 29 | 1064 |
| QIZ08096.1 | 0.0 | 2 | 1042 | 7 | 1057 |
| QWI08562.1 | 0.0 | 2 | 958 | 7 | 978 |
| QZD54715.1 | 7.26e-207 | 33 | 907 | 33 | 925 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3CLW_A | 2.78e-29 | 46 | 522 | 10 | 501 | Crystalstructure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_B Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_C Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_D Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_E Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343],3CLW_F Crystal structure of conserved exported protein from Bacteroides fragilis [Bacteroides fragilis NCTC 9343] |
| 4CKQ_A | 7.09e-16 | 158 | 516 | 82 | 409 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQ9_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQB_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQC_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQD_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQE_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
| 4UQA_A | 3.96e-15 | 158 | 516 | 82 | 409 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
| 5A6L_A | 3.96e-15 | 158 | 516 | 82 | 409 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],5A6M_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus ATCC 27405] |
| 4FMV_A | 1.03e-10 | 163 | 516 | 76 | 385 | CrystalStructure Analysis of a GH30 Endoxylanase from Clostridium papyrosolvens C71 [Ruminiclostridium papyrosolvens DSM 2782] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A0A401ETL2 | 1.16e-91 | 20 | 909 | 15 | 1064 | Exo-beta-1,6-galactobiohydrolase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=bl1,6Gal PE=1 SV=1 |
| Q76FP5 | 5.53e-56 | 41 | 520 | 21 | 476 | Endo-beta-1,6-galactanase OS=Hypocrea rufa OX=5547 GN=6GAL PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
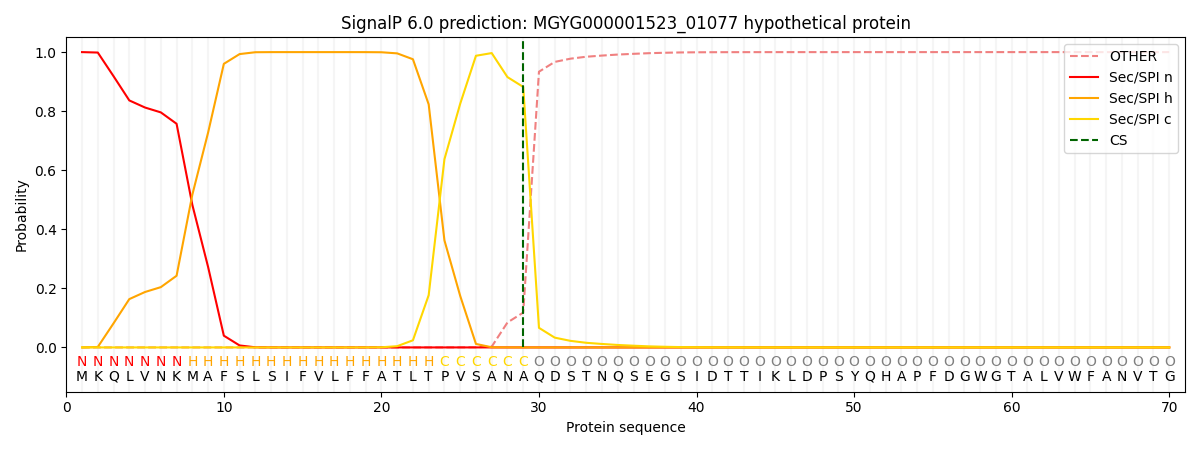
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000338 | 0.998846 | 0.000193 | 0.000230 | 0.000191 | 0.000176 |