You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001523_03047
You are here: Home > Sequence: MGYG000001523_03047
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
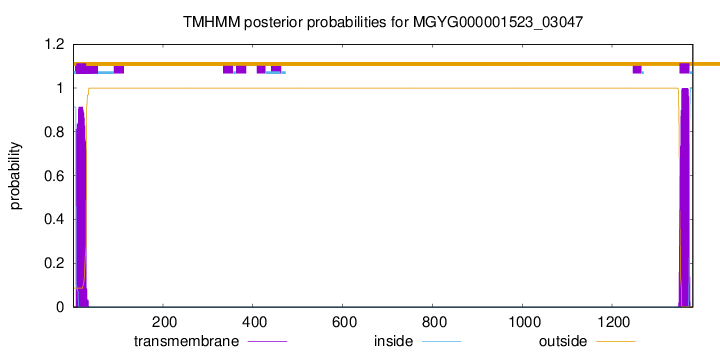
TMHMM annotations
Basic Information help
| Species | Gracilibacillus massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Bacillales_D; Amphibacillaceae; Gracilibacillus; Gracilibacillus massiliensis | |||||||||||
| CAZyme ID | MGYG000001523_03047 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 580764; End: 584906 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 99 | 293 | 1.3e-24 | 0.7592592592592593 |
| CBM6 | 1011 | 1145 | 9.6e-17 | 0.9130434782608695 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK15098 | PRK15098 | 1.35e-34 | 100 | 988 | 103 | 765 | beta-glucosidase BglX. |
| pfam01915 | Glyco_hydro_3_C | 1.01e-31 | 692 | 869 | 82 | 216 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| PLN03080 | PLN03080 | 5.44e-31 | 108 | 983 | 95 | 778 | Probable beta-xylosidase; Provisional |
| pfam14310 | Fn3-like | 2.79e-24 | 905 | 977 | 1 | 71 | Fibronectin type III-like domain. This domain has a fibronectin type III-like structure. It is often found in association with pfam00933 and pfam01915. Its function is unknown. |
| COG1472 | BglX | 6.40e-20 | 27 | 487 | 11 | 382 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AYA77556.1 | 0.0 | 32 | 1150 | 34 | 1149 |
| QKS56187.1 | 0.0 | 34 | 1150 | 35 | 1148 |
| AYQ75660.1 | 0.0 | 32 | 1152 | 33 | 1138 |
| QMV43973.1 | 0.0 | 39 | 1153 | 14 | 1123 |
| QOS79412.1 | 1.30e-243 | 51 | 1144 | 31 | 1121 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3AC0_A | 2.19e-28 | 118 | 989 | 64 | 837 | Crystalstructure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_B Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_C Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_D Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus] |
| 5YOT_A | 1.49e-26 | 693 | 985 | 467 | 753 | Isoprimeverose-producingenzyme from Aspergillus oryzae in complex with isoprimeverose [Aspergillus oryzae RIB40],5YOT_B Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose [Aspergillus oryzae RIB40],5YQS_A Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose [Aspergillus oryzae RIB40],5YQS_B Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose [Aspergillus oryzae RIB40] |
| 3ABZ_A | 1.79e-26 | 118 | 989 | 64 | 837 | Crystalstructure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus],3ABZ_B Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus],3ABZ_C Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus],3ABZ_D Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus] |
| 7EAP_A | 7.82e-26 | 693 | 985 | 467 | 753 | ChainA, Fn3_like domain-containing protein [Aspergillus oryzae RIB40] |
| 7MS2_A | 1.32e-25 | 693 | 992 | 396 | 672 | ChainA, Thermostable beta-glucosidase B [Acetivibrio thermocellus AD2],7MS2_B Chain B, Thermostable beta-glucosidase B [Acetivibrio thermocellus AD2] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P14002 | 7.24e-25 | 693 | 992 | 396 | 672 | Thermostable beta-glucosidase B OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=bglB PE=1 SV=2 |
| P07337 | 2.01e-24 | 118 | 989 | 64 | 837 | Beta-glucosidase OS=Kluyveromyces marxianus OX=4911 PE=3 SV=1 |
| A2QPK4 | 3.79e-24 | 698 | 991 | 472 | 753 | Probable beta-glucosidase D OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=bglD PE=3 SV=2 |
| D5EY15 | 1.41e-23 | 104 | 977 | 72 | 849 | Xylan 1,4-beta-xylosidase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=xyl3A PE=1 SV=1 |
| Q5AUW5 | 3.89e-23 | 702 | 981 | 534 | 801 | Probable beta-glucosidase D OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=bglD PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
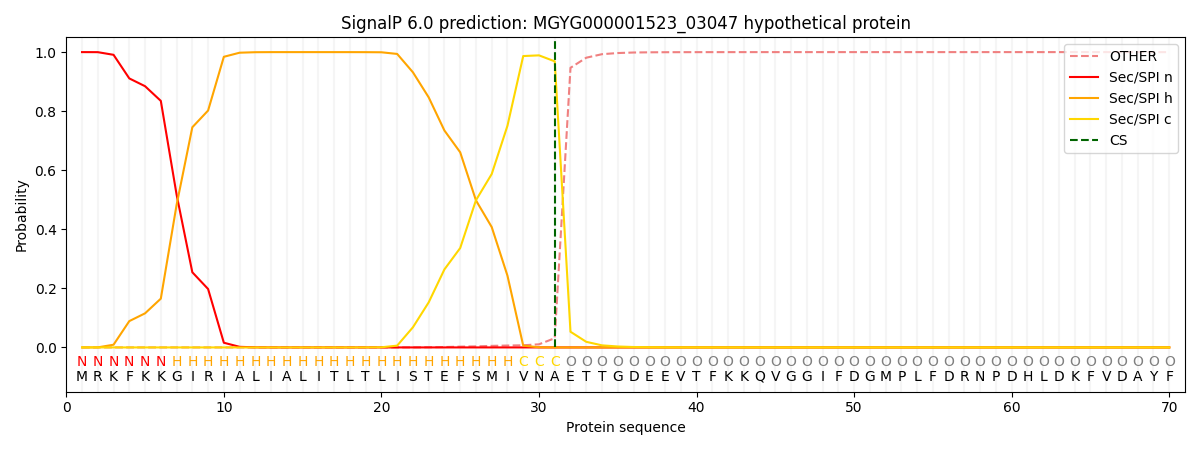
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000404 | 0.998718 | 0.000285 | 0.000210 | 0.000185 | 0.000167 |