You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001525_00862
You are here: Home > Sequence: MGYG000001525_00862
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
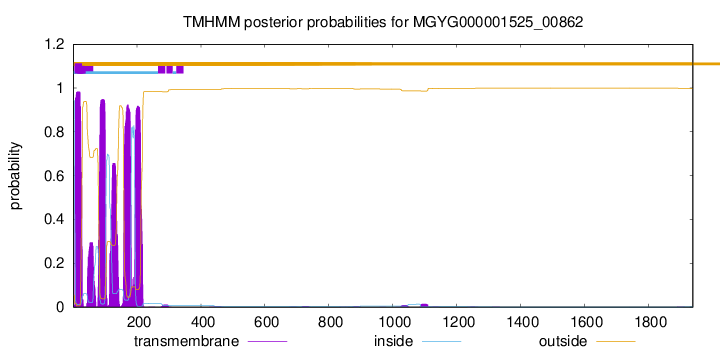
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A rubinfantis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A rubinfantis | |||||||||||
| CAZyme ID | MGYG000001525_00862 | |||||||||||
| CAZy Family | SLH | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 194490; End: 200312 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| SLH | 1824 | 1865 | 9.5e-16 | 0.9523809523809523 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033190 | inl_like_NEAT_1 | 3.19e-23 | 1756 | 1927 | 573 | 743 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| NF033189 | internalin_A | 1.05e-16 | 1364 | 1581 | 515 | 703 | class 1 internalin InlA. Internalins, as found in the intracellular human pathogen Listeria monocytogenes, are paralogous surface or secreted proteins with an N-terminal signal peptide, leucine-rich repeats, and usually a C-terminal LPXTG processing and cell surface anchoring site. See PMID:17764999 for a general discussion of internalins. Members of this family are internalin A (InlA), a class 1 (LPXTG-type) internalin. |
| NF033189 | internalin_A | 5.14e-13 | 1378 | 1552 | 596 | 761 | class 1 internalin InlA. Internalins, as found in the intracellular human pathogen Listeria monocytogenes, are paralogous surface or secreted proteins with an N-terminal signal peptide, leucine-rich repeats, and usually a C-terminal LPXTG processing and cell surface anchoring site. See PMID:17764999 for a general discussion of internalins. Members of this family are internalin A (InlA), a class 1 (LPXTG-type) internalin. |
| NF033190 | inl_like_NEAT_1 | 2.21e-12 | 1762 | 1875 | 640 | 753 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| NF033188 | internalin_H | 6.69e-12 | 1378 | 1541 | 374 | 525 | InlH/InlC2 family class 1 internalin. Internalins, as found in the intracellular human pathogen Listeria monocytogenes, are paralogous surface or secreted proteins with an N-terminal signal peptide, leucine-rich repeats, and usually a C-terminal LPXTG processing and cell surface anchoring site. See PMID:17764999 for a general discussion of internalins. Members of this family are internalin H (InlH), or internalin C2, two class 1 (LPXTG-type) internalins that are closely related, one apparently derived from the other through a recombination event. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QMV41393.1 | 2.11e-88 | 1619 | 1937 | 1917 | 2235 |
| QNK59241.1 | 4.07e-65 | 1623 | 1937 | 1149 | 1459 |
| QAV19712.1 | 4.66e-65 | 1621 | 1934 | 1207 | 1514 |
| BBI31951.1 | 6.66e-63 | 1622 | 1933 | 1224 | 1528 |
| QTH40658.1 | 1.44e-62 | 1615 | 1936 | 1374 | 1690 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6BT4_A | 3.05e-12 | 1765 | 1938 | 26 | 198 | Crystalstructure of the SLH domain of Sap from Bacillus anthracis in complex with a pyruvylated SCWP unit [Bacillus anthracis] |
| 3PYW_A | 3.11e-12 | 1765 | 1938 | 5 | 177 | Thestructure of the SLH domain from B. anthracis surface array protein at 1.8A [Bacillus anthracis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P38536 | 2.43e-29 | 1666 | 1939 | 1601 | 1858 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| P0DJ97 | 6.90e-28 | 1340 | 1583 | 429 | 661 | Putative Gly-rich membrane protein Bcell_0380 OS=Evansella cellulosilytica (strain ATCC 21833 / DSM 2522 / FERM P-1141 / JCM 9156 / N-4) OX=649639 GN=Bcell_0380 PE=4 SV=1 |
| P38535 | 1.95e-22 | 1765 | 1940 | 908 | 1085 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
| C6CRV0 | 1.32e-16 | 1733 | 1935 | 1264 | 1458 | Endo-1,4-beta-xylanase A OS=Paenibacillus sp. (strain JDR-2) OX=324057 GN=xynA1 PE=1 SV=1 |
| P0DJ98 | 2.63e-15 | 1341 | 1757 | 154 | 594 | Putative membrane protein Bcell_0381 OS=Evansella cellulosilytica (strain ATCC 21833 / DSM 2522 / FERM P-1141 / JCM 9156 / N-4) OX=649639 GN=Bcell_0381 PE=4 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
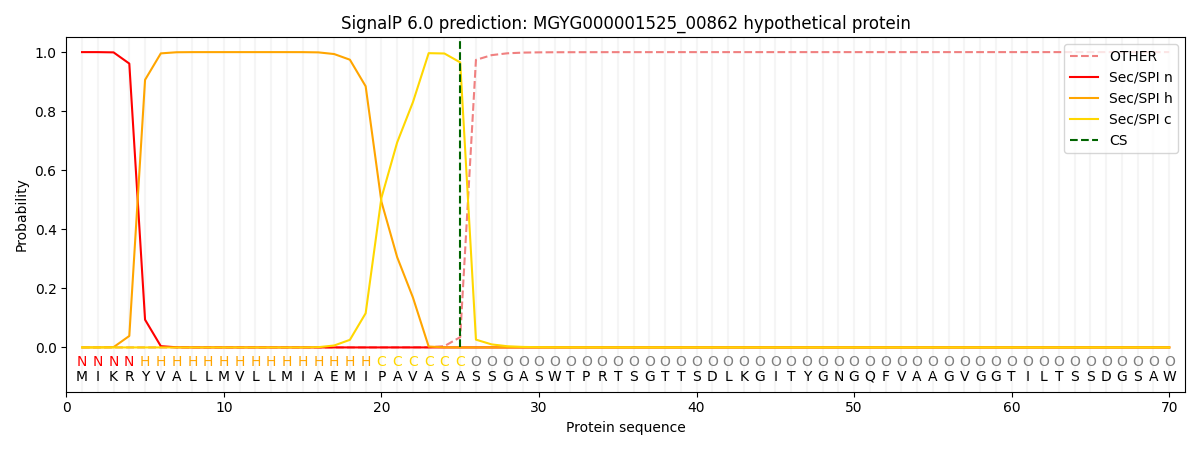
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000208 | 0.999101 | 0.000174 | 0.000178 | 0.000154 | 0.000146 |