You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001525_02473
You are here: Home > Sequence: MGYG000001525_02473
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
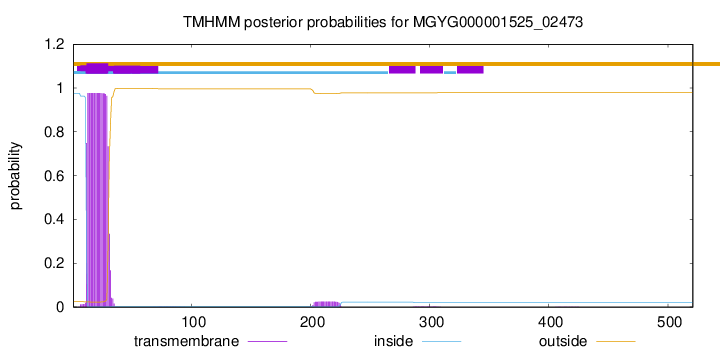
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A rubinfantis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A rubinfantis | |||||||||||
| CAZyme ID | MGYG000001525_02473 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 204558; End: 206123 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 41 | 352 | 3.5e-27 | 0.9527027027027027 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd02871 | GH18_chitinase_D-like | 7.49e-144 | 43 | 361 | 3 | 312 | GH18 domain of Chitinase D (ChiD). ChiD, a chitinase found in Bacillus circulans, hydrolyzes the 1,4-beta-linkages of N-acetylglucosamine in chitin and chitodextrins. The domain architecture of ChiD includes a catalytic glycosyl hydrolase family 18 (GH18) domain, a chitin-binding domain, and a fibronectin type III domain. The chitin-binding and fibronectin type III domains are located either N-terminal or C-terminal to the catalytic domain. This family includes exochitinase Chi36 from Bacillus cereus. |
| COG3469 | Chi1 | 3.32e-57 | 45 | 351 | 30 | 311 | Chitinase [Carbohydrate transport and metabolism]. |
| cd00598 | GH18_chitinase-like | 4.34e-23 | 43 | 252 | 1 | 195 | The GH18 (glycosyl hydrolase, family 18) type II chitinases hydrolyze chitin, an abundant polymer of beta-1,4-linked N-acetylglucosamine (GlcNAc) which is a major component of the cell wall of fungi and the exoskeleton of arthropods. Chitinases have been identified in viruses, bacteria, fungi, protozoan parasites, insects, and plants. The structure of the GH18 domain is an eight-stranded beta/alpha barrel with a pronounced active-site cleft at the C-terminal end of the beta-barrel. The GH18 family includes chitotriosidase, chitobiase, hevamine, zymocin-alpha, narbonin, SI-CLP (stabilin-1 interacting chitinase-like protein), IDGF (imaginal disc growth factor), CFLE (cortical fragment-lytic enzyme) spore hydrolase, the type III and type V plant chitinases, the endo-beta-N-acetylglucosaminidases, and the chitolectins. The GH85 (glycosyl hydrolase, family 85) ENGases (endo-beta-N-acetylglucosaminidases) are closely related to the GH18 chitinases and are included in this alignment model. |
| pfam00704 | Glyco_hydro_18 | 4.81e-21 | 44 | 347 | 6 | 307 | Glycosyl hydrolases family 18. |
| smart00636 | Glyco_18 | 4.31e-19 | 42 | 350 | 1 | 289 | Glyco_18 domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QTH43347.1 | 3.55e-233 | 15 | 521 | 17 | 602 |
| AZS15636.1 | 4.21e-181 | 5 | 521 | 4 | 516 |
| ATB35645.1 | 9.91e-180 | 37 | 365 | 282 | 610 |
| SYX83767.1 | 1.01e-177 | 13 | 521 | 12 | 612 |
| ADO73088.1 | 1.72e-177 | 39 | 363 | 281 | 605 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3N11_A | 1.84e-137 | 35 | 365 | 1 | 329 | Crystalstricture of wild-type chitinase from Bacillus cereus NCTU2 [Bacillus cereus],3N12_A Crystal stricture of chitinase in complex with zinc atoms from Bacillus cereus NCTU2 [Bacillus cereus] |
| 3N18_A | 5.24e-137 | 35 | 365 | 1 | 329 | Crystalstricture of E145G/Y227F chitinase in complex with NAG from Bacillus cereus NCTU2 [Bacillus cereus],3N1A_A Crystal stricture of E145G/Y227F chitinase in complex with cyclo-(L-His-L-Pro) from Bacillus cereus NCTU2 [Bacillus cereus] |
| 3N15_A | 5.24e-137 | 35 | 365 | 1 | 329 | Crystalstricture of E145Q chitinase in complex with NAG from Bacillus cereus NCTU2 [Bacillus cereus] |
| 5KZ6_A | 5.82e-137 | 35 | 365 | 4 | 332 | 1.25Angstrom Crystal Structure of Chitinase from Bacillus anthracis. [Bacillus anthracis],5KZ6_B 1.25 Angstrom Crystal Structure of Chitinase from Bacillus anthracis. [Bacillus anthracis] |
| 3N17_A | 2.11e-136 | 35 | 365 | 1 | 329 | Crystalstricture of E145Q/Y227F chitinase in complex with NAG from Bacillus cereus NCTU2 [Bacillus cereus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P27050 | 3.21e-155 | 36 | 367 | 185 | 516 | Chitinase D OS=Niallia circulans OX=1397 GN=chiD PE=1 SV=4 |
| Q05638 | 2.37e-90 | 25 | 362 | 248 | 596 | Exochitinase 1 OS=Streptomyces olivaceoviridis OX=1921 GN=chi01 PE=1 SV=1 |
| D4AVJ0 | 1.07e-84 | 59 | 365 | 17 | 331 | Probable class II chitinase ARB_00204 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_00204 PE=1 SV=2 |
| A5FB63 | 1.54e-58 | 43 | 362 | 1144 | 1480 | Chitinase ChiA OS=Flavobacterium johnsoniae (strain ATCC 17061 / DSM 2064 / JCM 8514 / NBRC 14942 / NCIMB 11054 / UW101) OX=376686 GN=chiA PE=1 SV=1 |
| E9F7R6 | 1.36e-42 | 32 | 362 | 36 | 345 | Endochitinase 4 OS=Metarhizium robertsii (strain ARSEF 23 / ATCC MYA-3075) OX=655844 GN=chi4 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
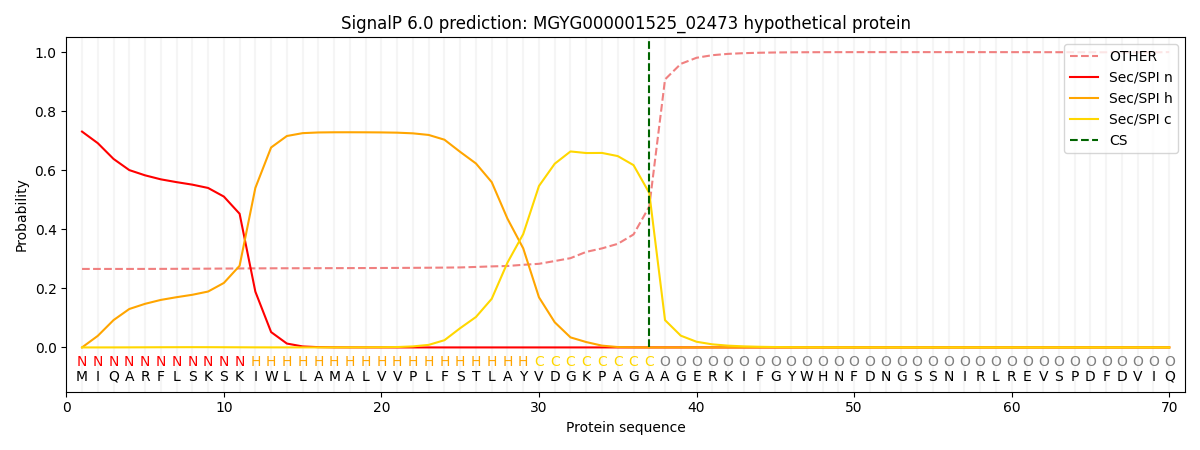
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.272847 | 0.721990 | 0.004009 | 0.000442 | 0.000303 | 0.000393 |