You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001525_04732
You are here: Home > Sequence: MGYG000001525_04732
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A rubinfantis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A rubinfantis | |||||||||||
| CAZyme ID | MGYG000001525_04732 | |||||||||||
| CAZy Family | GH27 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1305423; End: 1308506 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH27 | 437 | 686 | 4.2e-18 | 0.9519650655021834 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14791 | GH36 | 2.45e-22 | 309 | 614 | 3 | 299 | glycosyl hydrolase family 36 (GH36). GH36 enzymes occur in prokaryotes, eukaryotes, and archaea with a wide range of hydrolytic activities, including alpha-galactosidase, alpha-N-acetylgalactosaminidase, stachyose synthase, and raffinose synthase. All GH36 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. GH36 members are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
| cd14792 | GH27 | 1.54e-21 | 309 | 555 | 2 | 217 | glycosyl hydrolase family 27 (GH27). GH27 enzymes occur in eukaryotes, prokaryotes, and archaea with a wide range of hydrolytic activities, including alpha-glucosidase (glucoamylase and sucrase-isomaltase), alpha-N-acetylgalactosaminidase, and 3-alpha-isomalto-dextranase. All GH27 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. GH27 members are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
| pfam00754 | F5_F8_type_C | 3.92e-11 | 728 | 845 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| COG3345 | GalA | 1.88e-06 | 288 | 466 | 271 | 462 | Alpha-galactosidase [Carbohydrate transport and metabolism]. |
| cd14790 | GH_D | 3.85e-06 | 309 | 555 | 2 | 205 | Glycoside hydrolases, clan D. This group of glycosyl hydrolase families is comprised of glycosyl hydrolase family 31 (GH31), family 36 (GH36), and family 27 (GH27). These structurally and mechanistically related protein families are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. They have a wide range of functions including alpha-glucosidase, alpha-xylosidase, 6-alpha-glucosyltransferase, 3-alpha-isomaltosyltransferase, alpha-N-acetylgalactosaminidase, stachyose synthase, raffinose synthase, and alpha-1,4-glucan lyase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AYQ71140.1 | 0.0 | 35 | 1017 | 42 | 1025 |
| ANS75585.1 | 0.0 | 35 | 1016 | 41 | 1027 |
| AYN29307.1 | 0.0 | 30 | 1019 | 20 | 969 |
| AJQ98020.1 | 0.0 | 40 | 1019 | 34 | 983 |
| QQE42004.1 | 0.0 | 40 | 1019 | 34 | 983 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2RV9_A | 1.75e-06 | 722 | 848 | 7 | 135 | Solutionstructure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
| 4ZXE_A | 1.79e-06 | 722 | 848 | 8 | 136 | X-raycrystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis],4ZXE_B X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis],4ZXE_C X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis] |
| 4ZY9_A | 3.32e-06 | 722 | 848 | 8 | 136 | X-raycrystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZY9_B X-ray crystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
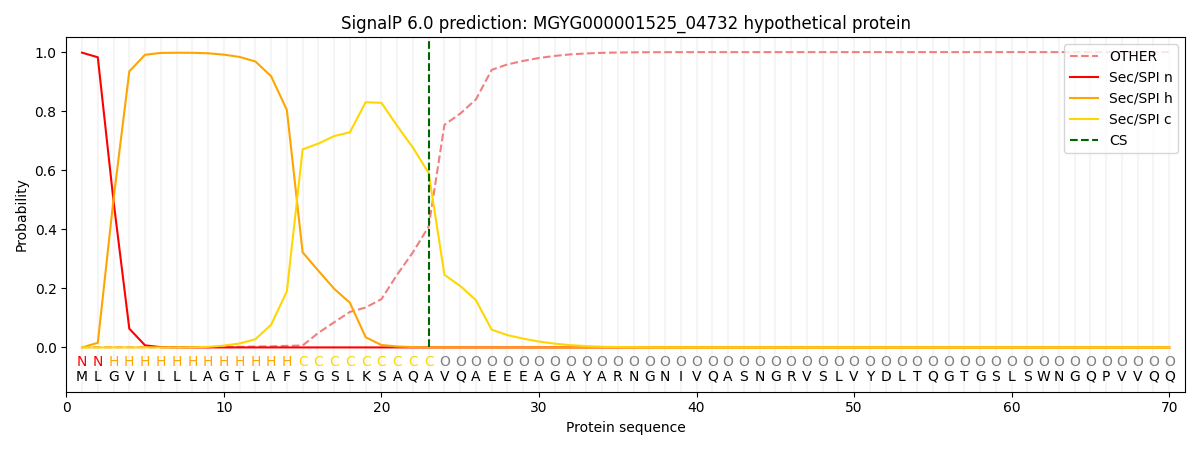
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001815 | 0.995831 | 0.001636 | 0.000278 | 0.000213 | 0.000181 |
