You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001525_04836
You are here: Home > Sequence: MGYG000001525_04836
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A rubinfantis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A rubinfantis | |||||||||||
| CAZyme ID | MGYG000001525_04836 | |||||||||||
| CAZy Family | GH11 | |||||||||||
| CAZyme Description | Endo-1,4-beta-xylanase A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1427939; End: 1428571 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH11 | 30 | 208 | 2.4e-71 | 0.9830508474576272 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00457 | Glyco_hydro_11 | 8.26e-88 | 30 | 206 | 3 | 175 | Glycosyl hydrolases family 11. |
| PRK06215 | PRK06215 | 0.002 | 1 | 134 | 9 | 167 | hypothetical protein; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AKJ70928.1 | 1.99e-146 | 1 | 210 | 1 | 210 |
| AAB72117.1 | 3.17e-143 | 1 | 210 | 1 | 210 |
| ALP35915.1 | 6.35e-141 | 2 | 210 | 3 | 211 |
| QOS78966.1 | 6.35e-141 | 2 | 210 | 3 | 211 |
| AYB44081.1 | 9.02e-141 | 2 | 210 | 3 | 211 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7KV0_A | 7.11e-129 | 28 | 210 | 4 | 186 | ChainA, Endo-1,4-beta-xylanase [Paenibacillus xylanivorans],7KV0_B Chain B, Endo-1,4-beta-xylanase [Paenibacillus xylanivorans] |
| 1XXN_A | 6.04e-123 | 27 | 210 | 1 | 185 | Crystalstructure of a mesophilic xylanase A from Bacillus subtilis 1A1 [Bacillus subtilis],2DCY_A Crystal structure of Bacillus subtilis family-11 xylanase [Bacillus subtilis],2DCY_B Crystal structure of Bacillus subtilis family-11 xylanase [Bacillus subtilis],2DCY_C Crystal structure of Bacillus subtilis family-11 xylanase [Bacillus subtilis],2DCY_D Crystal structure of Bacillus subtilis family-11 xylanase [Bacillus subtilis],2DCY_E Crystal structure of Bacillus subtilis family-11 xylanase [Bacillus subtilis] |
| 1BVV_A | 1.73e-122 | 27 | 210 | 1 | 185 | ChainA, ENDO-1,4-BETA-XYLANASE [Niallia circulans],1XNB_A Chain A, XYLANASE [Niallia circulans],2B42_B Crystal structure of the Triticum xylanse inhibitor-I in complex with bacillus subtilis xylanase [Bacillus subtilis],3HD8_B Crystal structure of the Triticum aestivum xylanase inhibitor-IIA in complex with bacillus subtilis xylanase [Bacillus subtilis],3HD8_D Crystal structure of the Triticum aestivum xylanase inhibitor-IIA in complex with bacillus subtilis xylanase [Bacillus subtilis] |
| 2QZ3_A | 4.96e-122 | 27 | 210 | 1 | 185 | ChainA, Endo-1,4-beta-xylanase A [Bacillus subtilis],2QZ3_B Chain B, Endo-1,4-beta-xylanase A [Bacillus subtilis],2Z79_A Chain A, Endo-1,4-beta-xylanase A [Bacillus subtilis],2Z79_B Chain B, Endo-1,4-beta-xylanase A [Bacillus subtilis] |
| 1HV0_A | 7.04e-122 | 27 | 210 | 1 | 185 | ChainA, ENDO-1,4-BETA-XYLANASE [Niallia circulans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P45705 | 6.33e-144 | 1 | 210 | 1 | 210 | Endo-1,4-beta-xylanase A OS=Geobacillus stearothermophilus OX=1422 GN=xynA PE=3 SV=2 |
| V9TXH2 | 5.74e-131 | 2 | 210 | 3 | 210 | Endo-1,4-beta-xylanase Xyn11E OS=Paenibacillus barcinonensis OX=198119 GN=xyn11E PE=1 SV=1 |
| P18429 | 1.44e-127 | 2 | 210 | 3 | 213 | Endo-1,4-beta-xylanase A OS=Bacillus subtilis (strain 168) OX=224308 GN=xynA PE=1 SV=1 |
| P09850 | 4.12e-127 | 2 | 210 | 3 | 213 | Endo-1,4-beta-xylanase OS=Niallia circulans OX=1397 GN=xlnA PE=1 SV=1 |
| Q9RI72 | 3.85e-80 | 24 | 209 | 55 | 239 | Endo-1,4-beta-xylanase C OS=Streptomyces coelicolor (strain ATCC BAA-471 / A3(2) / M145) OX=100226 GN=xlnC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
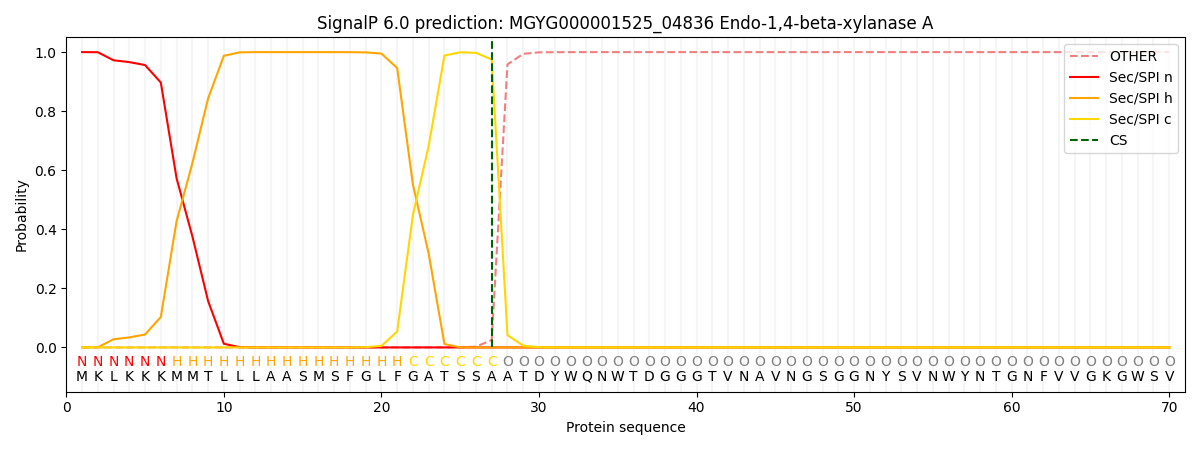
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000208 | 0.999146 | 0.000156 | 0.000169 | 0.000145 | 0.000134 |
