You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001526_01775
You are here: Home > Sequence: MGYG000001526_01775
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
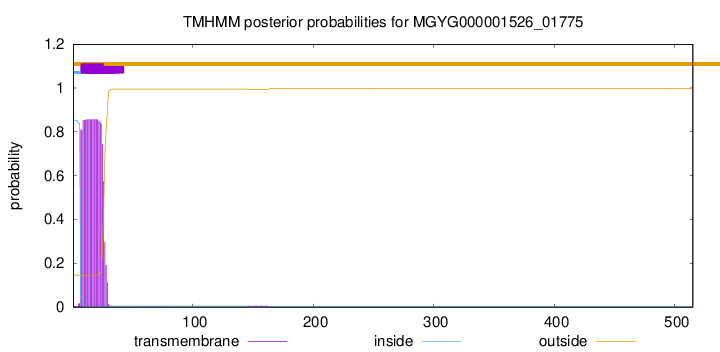
TMHMM annotations
Basic Information help
| Species | Paenibacillus_A senegalimassiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Paenibacillales; Paenibacillaceae; Paenibacillus_A; Paenibacillus_A senegalimassiliensis | |||||||||||
| CAZyme ID | MGYG000001526_01775 | |||||||||||
| CAZy Family | PL9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 640046; End: 641593 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 34 | 408 | 7.6e-150 | 0.9891304347826086 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13229 | Beta_helix | 5.66e-06 | 226 | 397 | 7 | 156 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 1.14e-05 | 291 | 423 | 20 | 137 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam07602 | DUF1565 | 1.34e-05 | 41 | 245 | 1 | 201 | Protein of unknown function (DUF1565). These proteins share a region of homology in their N termini, and are found in several phylogenetically diverse bacteria and in the archaeon Methanosarcina acetivorans. Some of these proteins also contain characterized domains such as pfam00395 and pfam03422. |
| pfam05048 | NosD | 3.83e-05 | 240 | 423 | 2 | 182 | Periplasmic copper-binding protein (NosD). NosD is a periplasmic protein which is thought to insert copper into the exported reductase apoenzyme (NosZ). This region forms a parallel beta helix domain. |
| pfam13229 | Beta_helix | 1.70e-04 | 150 | 369 | 2 | 157 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QLG40737.1 | 8.33e-299 | 1 | 515 | 1 | 515 |
| QOS78982.1 | 2.77e-297 | 1 | 515 | 1 | 515 |
| ASR46616.1 | 3.76e-294 | 12 | 515 | 12 | 511 |
| AET61906.1 | 1.19e-290 | 12 | 515 | 12 | 511 |
| AOK89169.1 | 9.28e-288 | 3 | 515 | 3 | 511 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P94576 | 1.15e-95 | 4 | 482 | 1 | 463 | Uncharacterized protein YwoF OS=Bacillus subtilis (strain 168) OX=224308 GN=ywoF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
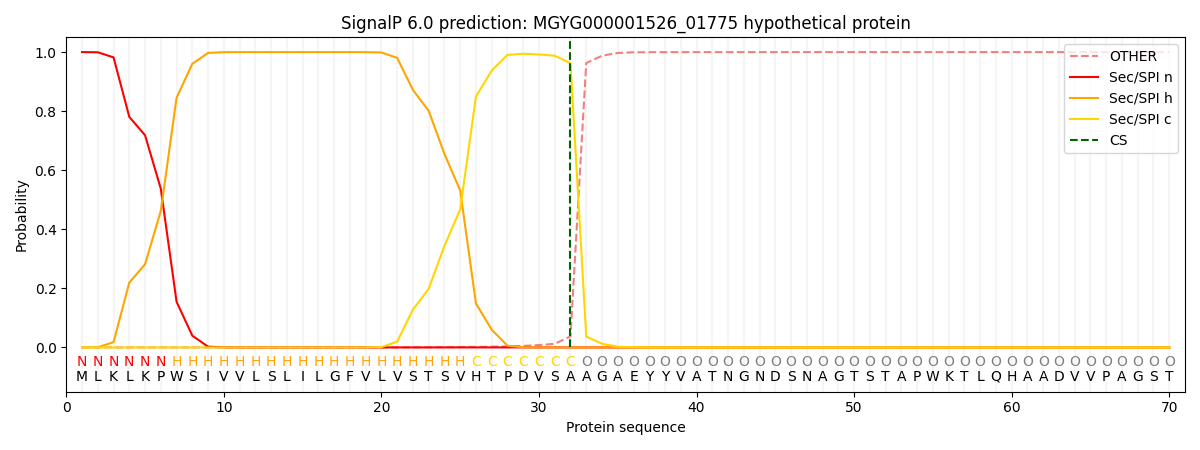
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000248 | 0.999101 | 0.000181 | 0.000161 | 0.000143 | 0.000132 |