You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001528_02169
You are here: Home > Sequence: MGYG000001528_02169
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
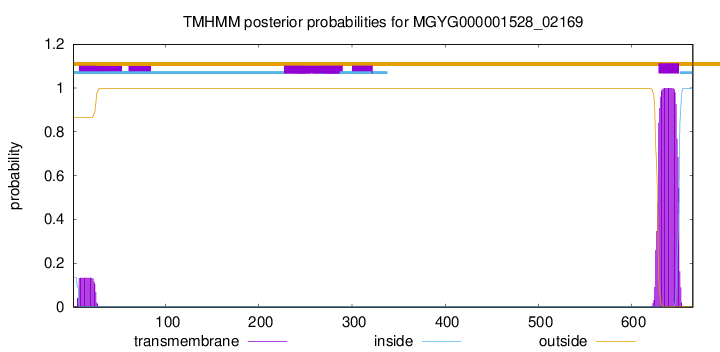
TMHMM annotations
Basic Information help
| Species | Mediterraneibacter massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Mediterraneibacter; Mediterraneibacter massiliensis | |||||||||||
| CAZyme ID | MGYG000001528_02169 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1355021; End: 1357021 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 1.05e-23 | 115 | 348 | 38 | 244 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| cd08547 | Type_II_cohesin | 7.11e-04 | 408 | 503 | 5 | 116 | Type II cohesin domain, interaction partner of dockerin. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type II cohesins; their interactions with dockerin mediate attachment of the cellulosome complex to the bacterial cell wall. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRV21758.1 | 3.96e-108 | 41 | 365 | 117 | 523 |
| ADL03932.1 | 3.96e-108 | 41 | 365 | 117 | 523 |
| CBK77705.1 | 1.95e-103 | 41 | 360 | 120 | 540 |
| ASN95383.1 | 8.32e-96 | 41 | 365 | 118 | 599 |
| QRP39925.1 | 8.32e-96 | 41 | 365 | 118 | 599 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
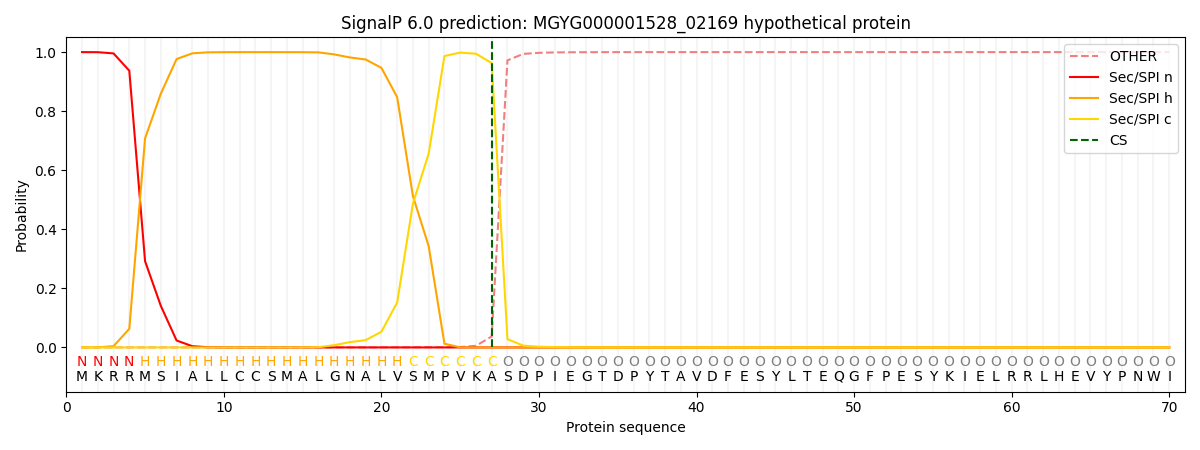
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000431 | 0.998721 | 0.000260 | 0.000235 | 0.000171 | 0.000142 |