You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001533_00214
You are here: Home > Sequence: MGYG000001533_00214
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Corynebacterium ammoniagenes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Corynebacterium; Corynebacterium ammoniagenes | |||||||||||
| CAZyme ID | MGYG000001533_00214 | |||||||||||
| CAZy Family | GT53 | |||||||||||
| CAZyme Description | putative arabinosyltransferase C | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 197352; End: 200747 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT53 | 21 | 1124 | 0 | 0.9952335557673975 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam04602 | Arabinose_trans | 0.0 | 207 | 666 | 1 | 459 | Mycobacterial cell wall arabinan synthesis protein. Arabinosyltransferase is involved in arabinogalactan (AG) biosynthesis pathway in mycobacteria. AG is a component of the macromolecular assembly of the mycolyl-AG-peptidoglycan complex of the cell wall. This enzyme has important clinical applications as it is believed to be the target of the antimycobacterial drug Ethambutol. |
| pfam14896 | Arabino_trans_C | 3.40e-175 | 706 | 1124 | 1 | 384 | EmbC C-terminal domain. Arabinosyltransferase is involved in arabinogalactan (AG) biosynthesis pathway in mycobacteria. AG is a component of the macromolecular assembly of the mycolyl-AG-peptidoglycan complex of the cell wall. This enzyme has important clinical applications as it is believed to be the target of the antimycobacterial drug Ethambutol. This domain represents the C-terminal extracellular domain that is likely to bind to carbohydrate. |
| pfam17689 | Arabino_trans_N | 2.59e-51 | 45 | 203 | 1 | 155 | Arabinosyltransferase concanavalin like domain. Arabinosyltransferase is involved in arabinogalactan (AG) biosynthesis pathway in mycobacteria. AG is a component of the macromolecular assembly of the mycolyl-AG-peptidoglycan complex of the cell wall. This enzyme has important clinical applications as it is believed to be the target of the antimycobacterial drug Ethambutol. |
| COG1807 | ArnT | 0.001 | 257 | 670 | 15 | 411 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| COG0814 | SdaC | 0.004 | 518 | 708 | 48 | 252 | Amino acid permease [Amino acid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| APT81579.1 | 0.0 | 1 | 1131 | 1 | 1131 |
| AQS72703.1 | 0.0 | 1 | 1131 | 1 | 1131 |
| AMJ43631.1 | 0.0 | 1 | 1131 | 1 | 1131 |
| AQX72304.1 | 0.0 | 1 | 1131 | 1 | 1131 |
| AQX70078.1 | 0.0 | 1 | 1131 | 1 | 1131 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7BVE_A | 6.75e-264 | 22 | 1125 | 16 | 1073 | Cryo-EMstructure of Mycobacterium smegmatis arabinosyltransferase EmbC2-AcpM2 in complex with ethambutol [Mycolicibacterium smegmatis MC2 155],7BVE_B Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbC2-AcpM2 in complex with ethambutol [Mycolicibacterium smegmatis MC2 155] |
| 7BVH_A | 1.67e-263 | 22 | 1125 | 29 | 1086 | Crystalstructure of arabinosyltransferase EmbC2-AcpM2 complex from Mycobacterium smegmatis complexed with di-arabinose [Mycolicibacterium smegmatis MC2 155],7BVH_B Crystal structure of arabinosyltransferase EmbC2-AcpM2 complex from Mycobacterium smegmatis complexed with di-arabinose [Mycolicibacterium smegmatis MC2 155] |
| 7BWR_A | 1.98e-233 | 22 | 1126 | 27 | 1081 | Mycobacteriumsmegmatis arabinosyltransferase complex EmbB2-AcpM2 in substrate DPA bound asymmetric 'active state' [Mycolicibacterium smegmatis MC2 155],7BWR_B Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in substrate DPA bound asymmetric 'active state' [Mycolicibacterium smegmatis MC2 155],7BX8_A Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in symmetric 'resting state' [Mycolicibacterium smegmatis MC2 155],7BX8_B Mycobacterium smegmatis arabinosyltransferase complex EmbB2-AcpM2 in symmetric 'resting state' [Mycolicibacterium smegmatis MC2 155] |
| 6X0O_A | 3.20e-233 | 22 | 1126 | 27 | 1081 | Single-ParticleCryo-EM Structure of Arabinosyltransferase EmbB from Mycobacterium smegmatis [Mycolicibacterium smegmatis MC2 155] |
| 7BVC_B | 3.40e-233 | 22 | 1126 | 27 | 1081 | Cryo-EMstructure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol [Mycolicibacterium smegmatis MC2 155],7BVG_B Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with di-arabinose. [Mycolicibacterium smegmatis MC2 155] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q50393 | 1.90e-260 | 22 | 1125 | 16 | 1073 | Probable arabinosyltransferase C OS=Mycolicibacterium smegmatis OX=1772 GN=embC PE=3 SV=2 |
| Q7TVN4 | 5.74e-246 | 9 | 1124 | 14 | 1092 | Probable arabinosyltransferase C OS=Mycobacterium bovis (strain ATCC BAA-935 / AF2122/97) OX=233413 GN=embC PE=3 SV=1 |
| P9WNL5 | 3.21e-245 | 9 | 1124 | 14 | 1092 | Probable arabinosyltransferase C OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=embC PE=1 SV=1 |
| P9WNL4 | 4.52e-245 | 9 | 1124 | 14 | 1092 | Probable arabinosyltransferase C OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=embC PE=3 SV=1 |
| Q9CDA7 | 1.06e-242 | 22 | 1124 | 12 | 1069 | Probable arabinosyltransferase C OS=Mycobacterium leprae (strain TN) OX=272631 GN=embC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000029 | 0.000008 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
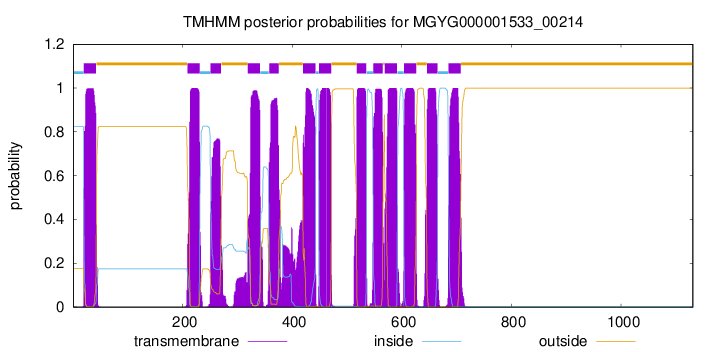
| start | end |
|---|---|
| 20 | 42 |
| 209 | 231 |
| 251 | 270 |
| 319 | 341 |
| 358 | 375 |
| 420 | 442 |
| 449 | 471 |
| 518 | 535 |
| 548 | 565 |
| 569 | 591 |
| 604 | 626 |
| 646 | 665 |
| 685 | 707 |
