You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001533_01318
You are here: Home > Sequence: MGYG000001533_01318
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Corynebacterium ammoniagenes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Mycobacteriales; Mycobacteriaceae; Corynebacterium; Corynebacterium ammoniagenes | |||||||||||
| CAZyme ID | MGYG000001533_01318 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1396998; End: 1398065 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 88 | 218 | 7.5e-25 | 0.5903083700440529 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3509 | LpqC | 1.01e-37 | 88 | 332 | 46 | 309 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| TIGR01840 | esterase_phb | 2.30e-12 | 93 | 213 | 3 | 131 | esterase, PHB depolymerase family. This model describes a subfamily among lipases of the ab-hydrolase family. This subfamily includes bacterial depolymerases for poly(3-hydroxybutyrate) (PHB) and related polyhydroxyalkanoates (PHA), as well as acetyl xylan esterases, feruloyl esterases, and others from fungi. [Fatty acid and phospholipid metabolism, Degradation] |
| COG4099 | COG4099 | 2.95e-08 | 90 | 238 | 175 | 329 | Predicted peptidase [General function prediction only]. |
| pfam00756 | Esterase | 7.14e-08 | 89 | 209 | 8 | 142 | Putative esterase. This family contains Esterase D. However it is not clear if all members of the family have the same function. This family is related to the pfam00135 family. |
| COG0400 | YpfH | 1.09e-07 | 101 | 252 | 14 | 174 | Predicted esterase [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUX40426.1 | 1.13e-22 | 65 | 238 | 232 | 398 |
| AUX29931.1 | 2.14e-22 | 65 | 271 | 236 | 425 |
| ALO15718.1 | 3.24e-22 | 72 | 328 | 18 | 287 |
| CAN90905.1 | 7.36e-22 | 65 | 243 | 237 | 408 |
| AGP34778.1 | 3.44e-21 | 65 | 243 | 239 | 410 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3WYD_A | 4.14e-07 | 80 | 209 | 12 | 141 | C-terminalesterase domain of LC-Est1 [uncultured organism],3WYD_B C-terminal esterase domain of LC-Est1 [uncultured organism] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A1CC33 | 3.64e-19 | 70 | 327 | 26 | 269 | Probable feruloyl esterase C OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=faeC-2 PE=3 SV=1 |
| Q9Y871 | 1.89e-15 | 89 | 302 | 292 | 519 | Feruloyl esterase B OS=Piromyces equi OX=99929 GN=ESTA PE=2 SV=1 |
| Q5B2G3 | 4.27e-15 | 69 | 327 | 23 | 267 | Feruloyl esterase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=faeC PE=1 SV=1 |
| A2QYU7 | 3.16e-13 | 69 | 250 | 23 | 205 | Probable feruloyl esterase C OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=faeC PE=3 SV=1 |
| B8YG19 | 7.47e-10 | 87 | 301 | 56 | 263 | Bifunctional acetylxylan esterase/xylanase XynS20E OS=Neocallimastix patriciarum OX=4758 GN=xynS20E PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
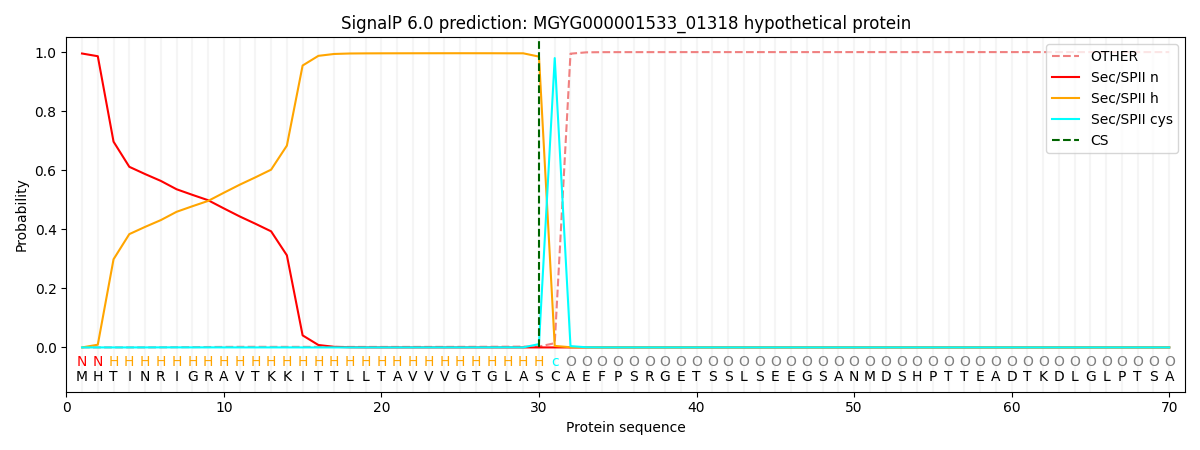
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000036 | 0.001783 | 0.998189 | 0.000003 | 0.000002 | 0.000001 |
