You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001537_00728
You are here: Home > Sequence: MGYG000001537_00728
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
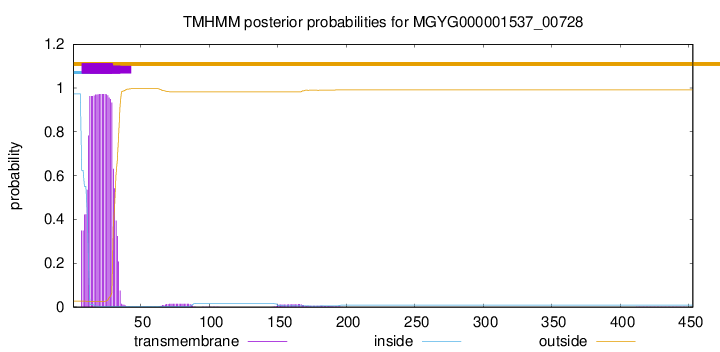
TMHMM annotations
Basic Information help
| Species | Cellulomonas timonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Cellulomonadaceae; Cellulomonas; Cellulomonas timonensis | |||||||||||
| CAZyme ID | MGYG000001537_00728 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 326842; End: 328203 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM2 | 351 | 450 | 2.5e-33 | 0.9900990099009901 |
| CE1 | 55 | 251 | 1.8e-19 | 0.7929515418502202 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR01840 | esterase_phb | 6.14e-38 | 56 | 269 | 1 | 211 | esterase, PHB depolymerase family. This model describes a subfamily among lipases of the ab-hydrolase family. This subfamily includes bacterial depolymerases for poly(3-hydroxybutyrate) (PHB) and related polyhydroxyalkanoates (PHA), as well as acetyl xylan esterases, feruloyl esterases, and others from fungi. [Fatty acid and phospholipid metabolism, Degradation] |
| pfam00553 | CBM_2 | 6.54e-31 | 351 | 450 | 1 | 101 | Cellulose binding domain. Two tryptophan residues are involved in cellulose binding. Cellulose binding domain found in bacteria. |
| COG3509 | LpqC | 2.07e-25 | 19 | 299 | 13 | 277 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| smart00637 | CBD_II | 1.30e-22 | 357 | 449 | 1 | 92 | CBD_II domain. |
| pfam10503 | Esterase_phd | 1.61e-12 | 54 | 186 | 1 | 135 | Esterase PHB depolymerase. This family of proteins include acetyl xylan esterases (AXE), feruloyl esterases (FAE), and poly(3-hydroxybutyrate) (PHB) depolymerases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QVI65015.1 | 6.23e-212 | 10 | 450 | 7 | 454 |
| QVI61330.1 | 1.18e-201 | 5 | 450 | 10 | 480 |
| QCB93012.1 | 1.98e-195 | 5 | 450 | 11 | 470 |
| QWC15145.1 | 2.50e-191 | 5 | 453 | 10 | 483 |
| QLD25124.1 | 1.93e-185 | 19 | 450 | 11 | 448 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5X6S_A | 2.37e-74 | 40 | 310 | 3 | 273 | Acetylxylan esterase from Aspergillus awamori [Aspergillus awamori],5X6S_B Acetyl xylan esterase from Aspergillus awamori [Aspergillus awamori] |
| 1EXG_A | 5.21e-45 | 351 | 453 | 8 | 110 | ChainA, EXO-1,4-BETA-D-GLYCANASE [Cellulomonas fimi],1EXH_A Chain A, EXO-1,4-BETA-D-GLYCANASE [Cellulomonas fimi] |
| 6QFS_A | 7.26e-43 | 351 | 453 | 4 | 106 | ChainA, Exoglucanase/xylanase [Cellulomonas fimi],6QFS_B Chain B, Exoglucanase/xylanase [Cellulomonas fimi],6QFS_C Chain C, Exoglucanase/xylanase [Cellulomonas fimi],6QFS_D Chain D, Exoglucanase/xylanase [Cellulomonas fimi],6QFS_E Chain E, Exoglucanase/xylanase [Cellulomonas fimi],6QFS_F Chain F, Exoglucanase/xylanase [Cellulomonas fimi],6QFS_G Chain G, Exoglucanase/xylanase [Cellulomonas fimi],6QFS_H Chain H, Exoglucanase/xylanase [Cellulomonas fimi] |
| 3NDY_E | 1.82e-20 | 353 | 450 | 5 | 104 | Thestructure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans [Clostridium cellulovorans],3NDY_F The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans [Clostridium cellulovorans],3NDY_G The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans [Clostridium cellulovorans],3NDY_H The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans [Clostridium cellulovorans],3NDZ_E The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans bound to cellotriose [Clostridium cellulovorans],3NDZ_F The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans bound to cellotriose [Clostridium cellulovorans],3NDZ_G The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans bound to cellotriose [Clostridium cellulovorans],3NDZ_H The structure of the catalytic and carbohydrate binding domain of endoglucanase D from Clostridium cellulovorans bound to cellotriose [Clostridium cellulovorans] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8M9H9 | 8.40e-89 | 34 | 311 | 14 | 291 | Feruloyl esterase B OS=Talaromyces stipitatus (strain ATCC 10500 / CBS 375.48 / QM 6759 / NRRL 1006) OX=441959 GN=faeB PE=1 SV=1 |
| Q9HE18 | 3.20e-87 | 34 | 315 | 14 | 295 | Feruloyl esterase B OS=Talaromyces funiculosus OX=28572 GN=FAEB PE=1 SV=1 |
| Q4WBW4 | 4.81e-82 | 29 | 310 | 15 | 303 | Probable acetylxylan esterase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=axeA PE=3 SV=1 |
| A1DBP9 | 7.64e-80 | 29 | 310 | 15 | 303 | Probable acetylxylan esterase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=axeA PE=3 SV=1 |
| B8NBI2 | 9.14e-77 | 37 | 310 | 31 | 304 | Probable acetylxylan esterase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=axeA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
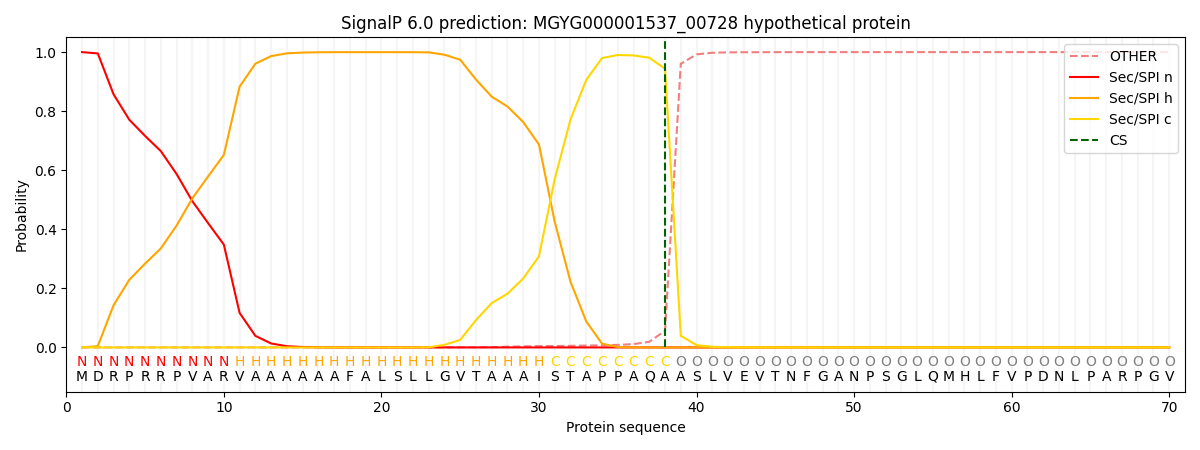
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000417 | 0.998764 | 0.000186 | 0.000267 | 0.000177 | 0.000153 |