You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001537_01011
You are here: Home > Sequence: MGYG000001537_01011
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
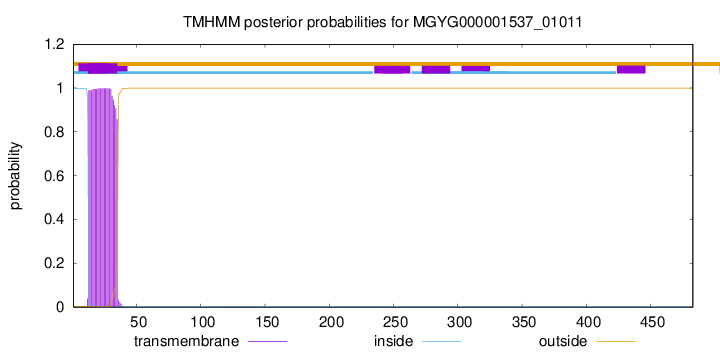
TMHMM annotations
Basic Information help
| Species | Cellulomonas timonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Cellulomonadaceae; Cellulomonas; Cellulomonas timonensis | |||||||||||
| CAZyme ID | MGYG000001537_01011 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | Exoglucanase/xylanase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 301268; End: 302719 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 41 | 344 | 3.8e-110 | 0.9801980198019802 |
| CBM2 | 381 | 480 | 5.5e-34 | 0.9900990099009901 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 7.27e-173 | 38 | 343 | 1 | 309 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 1.97e-134 | 80 | 342 | 1 | 263 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 3.69e-101 | 22 | 343 | 5 | 338 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00553 | CBM_2 | 1.44e-37 | 381 | 480 | 1 | 101 | Cellulose binding domain. Two tryptophan residues are involved in cellulose binding. Cellulose binding domain found in bacteria. |
| smart00637 | CBD_II | 4.12e-30 | 387 | 479 | 1 | 92 | CBD_II domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QVI64913.1 | 2.57e-221 | 51 | 480 | 52 | 483 |
| QHT56371.1 | 6.33e-221 | 3 | 483 | 33 | 521 |
| AAA56792.1 | 2.34e-219 | 3 | 482 | 11 | 484 |
| AEA30147.1 | 2.34e-219 | 3 | 482 | 11 | 484 |
| VEH29285.1 | 5.73e-219 | 3 | 482 | 36 | 509 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3CUF_A | 1.93e-170 | 35 | 347 | 1 | 313 | Cellulomonasfimi Xylanase/Cellulase Cex (Cf Xyn10A) in complex with cellobiose-like isofagomine [Cellulomonas fimi],3CUG_A Cellulomonas fimi Xylanase/Cellulase Cex (Cf Xyn10A) in complex with cellotetraose-like isofagomine [Cellulomonas fimi],3CUH_A Cellulomonas fimi Xylanase/Cellulase Cex (Cf Xyn10A) in complex with cellotriose-like isofagomine [Cellulomonas fimi],3CUI_A Cellulomonas fimi Xylanase/Cellulase Cex (Cf Xyn10A) in complex with sulfur substituted beta-1,4 xylotetraose [Cellulomonas fimi],3CUJ_A Cellulomonas fimi Xylanase/Cellulase Cex (Cf Xyn10A) in complex with sulfur substituted beta-1,4 xylopentaose. [Cellulomonas fimi] |
| 1EXP_A | 9.94e-170 | 35 | 345 | 1 | 311 | Beta-1,4-GlycanaseCex-Cd [Cellulomonas fimi],1FH7_A Crystal Structure Of The Xylanase Cex With Xylobiose- Derived Inhibitor Deoxynojirimycin [Cellulomonas fimi],1FH8_A Crystal Structure Of The Xylanase Cex With Xylobiose-derived Isofagomine Inhibitor [Cellulomonas fimi],1FH9_A Crystal Structure Of The Xylanase Cex With Xylobiose-derived Lactam Oxime Inhibitor [Cellulomonas fimi],1FHD_A Crystal Structure Of The Xylanase Cex With Xylobiose-derived Imidazole Inhibitor [Cellulomonas fimi],1J01_A Crystal Structure Of The Xylanase Cex With Xylobiose-Derived Inhibitor Isofagomine lactam [Cellulomonas fimi],2EXO_A Crystal Structure Of The Catalytic Domain Of The Beta-1,4- Glycanase Cex From Cellulomonas Fimi [Cellulomonas fimi],2XYL_A Cellulomonas Fimi XylanaseCELLULASE COMPLEXED WITH 2-Deoxy- 2-Fluoro-Xylobiose [Cellulomonas fimi] |
| 2HIS_A | 9.35e-168 | 35 | 345 | 1 | 311 | ChainA, CELLULOMONAS FIMI FAMILY 10 BETA-1,4-GLYCANASE [Cellulomonas fimi] |
| 1V6Y_A | 9.09e-134 | 34 | 347 | 1 | 315 | CrystalStructure Of chimeric Xylanase between Streptomyces Olivaceoviridis E-86 FXYN and Cellulomonas fimi Cex [Streptomyces olivaceoviridis] |
| 3WUF_A | 2.96e-124 | 37 | 346 | 4 | 312 | Themutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) from Streptomyces sp. 9 [Streptomyces sp.],3WUG_A The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) with xylobiose from Streptomyces sp. 9 [Streptomyces sp.] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P07986 | 1.73e-217 | 20 | 482 | 27 | 483 | Exoglucanase/xylanase OS=Cellulomonas fimi OX=1708 GN=cex PE=1 SV=1 |
| B4XVN1 | 5.27e-142 | 3 | 469 | 6 | 450 | Endo-1,4-beta-xylanase A OS=Streptomyces sp. OX=1931 GN=xynAS9 PE=1 SV=1 |
| O60206 | 9.81e-109 | 38 | 342 | 19 | 327 | Endo-1,4-beta-xylanase OS=Agaricus bisporus OX=5341 GN=xlnA PE=2 SV=1 |
| Q9HEZ1 | 7.94e-108 | 32 | 347 | 88 | 407 | Endo-1,4-beta-xylanase A OS=Phanerodontia chrysosporium OX=2822231 GN=xynA PE=1 SV=1 |
| B7SIW2 | 3.06e-99 | 38 | 343 | 84 | 394 | Endo-1,4-beta-xylanase C OS=Phanerodontia chrysosporium OX=2822231 GN=xynC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
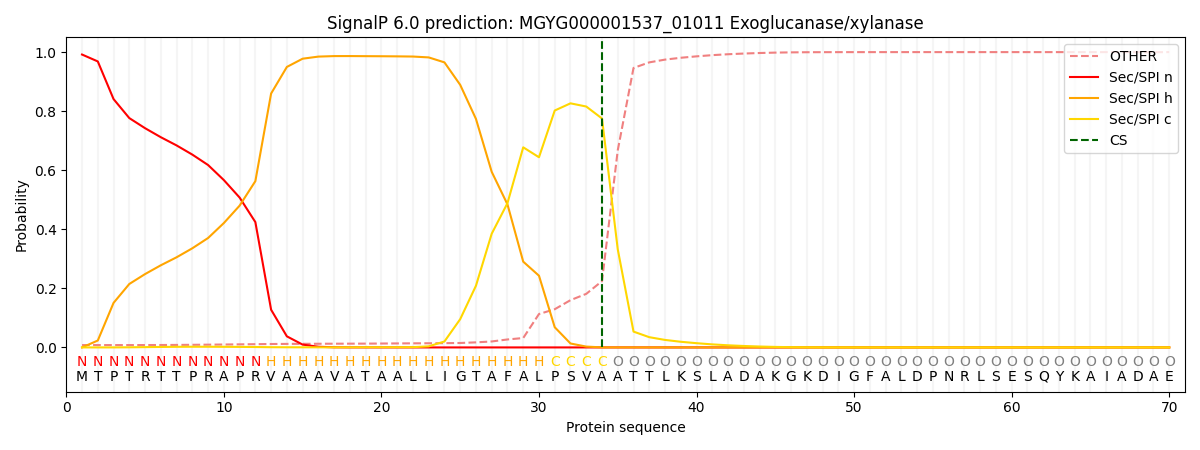
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.009607 | 0.988931 | 0.000433 | 0.000477 | 0.000277 | 0.000244 |