You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001537_01196
You are here: Home > Sequence: MGYG000001537_01196
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cellulomonas timonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Cellulomonadaceae; Cellulomonas; Cellulomonas timonensis | |||||||||||
| CAZyme ID | MGYG000001537_01196 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 84009; End: 85880 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 150 | 280 | 5.5e-18 | 0.37293729372937295 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4124 | ManB2 | 1.36e-09 | 164 | 272 | 198 | 300 | Beta-mannanase [Carbohydrate transport and metabolism]. |
| cd00063 | FN3 | 4.04e-05 | 536 | 611 | 9 | 82 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| smart00060 | FN3 | 0.005 | 528 | 610 | 1 | 81 | Fibronectin type 3 domain. One of three types of internal repeat within the plasma protein, fibronectin. The tenth fibronectin type III repeat contains a RGD cell recognition sequence in a flexible loop between 2 strands. Type III modules are present in both extracellular and intracellular proteins. |
| smart00089 | PKD | 0.006 | 341 | 424 | 6 | 77 | Repeats in polycystic kidney disease 1 (PKD1) and other proteins. Polycystic kidney disease 1 protein contains 14 repeats, present elsewhere such as in microbial collagenases. |
| pfam02156 | Glyco_hydro_26 | 0.007 | 162 | 228 | 170 | 230 | Glycosyl hydrolase family 26. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHT57318.1 | 7.90e-176 | 16 | 622 | 4 | 602 |
| AEE46219.1 | 2.29e-164 | 2 | 620 | 42 | 646 |
| VEH32108.1 | 2.29e-164 | 2 | 620 | 42 | 646 |
| ADG75638.1 | 9.38e-132 | 53 | 620 | 58 | 617 |
| QGQ20534.1 | 2.63e-122 | 53 | 620 | 1 | 563 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2BV9_A | 1.33e-26 | 50 | 330 | 7 | 284 | ChainA, Endoglucanase H [Acetivibrio thermocellus] |
| 2BVD_A | 2.89e-26 | 50 | 328 | 7 | 282 | ChainA, ENDOGLUCANASE H [Acetivibrio thermocellus] |
| 2V3G_A | 2.89e-26 | 50 | 328 | 7 | 282 | ChainA, Endoglucanase H [Acetivibrio thermocellus] |
| 2CIP_A | 7.10e-26 | 50 | 328 | 7 | 282 | ChainA, ENDOGLUCANASE H [Acetivibrio thermocellus] |
| 2CIT_A | 1.78e-25 | 50 | 328 | 7 | 282 | ChainA, ENDOGLUCANASE H [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16218 | 7.67e-24 | 50 | 332 | 29 | 308 | Endoglucanase H OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celH PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
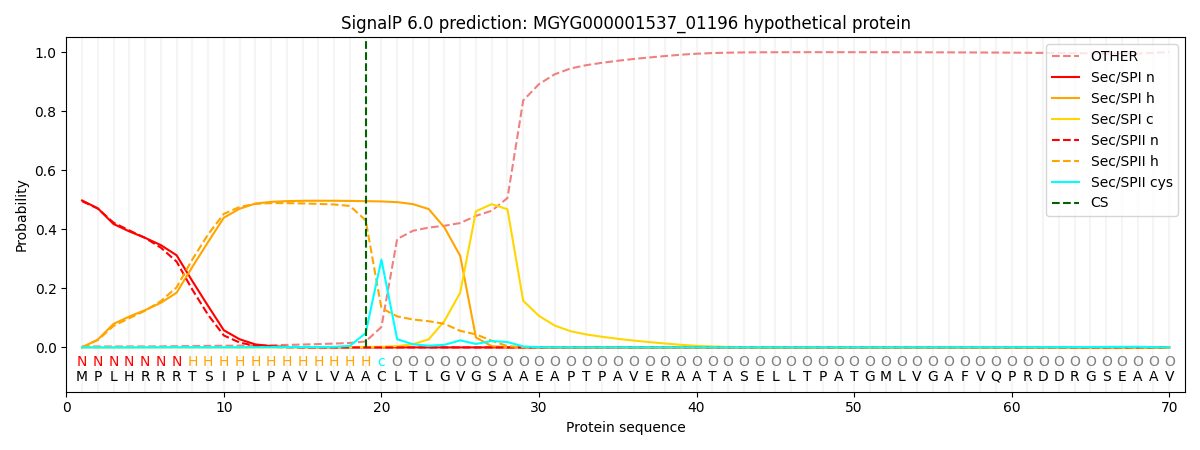
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.004974 | 0.484267 | 0.505123 | 0.004596 | 0.000686 | 0.000341 |
