You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001537_01427
You are here: Home > Sequence: MGYG000001537_01427
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
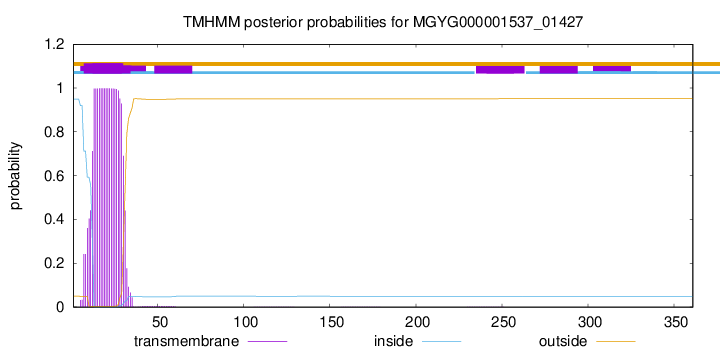
TMHMM annotations
Basic Information help
| Species | Cellulomonas timonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Cellulomonadaceae; Cellulomonas; Cellulomonas timonensis | |||||||||||
| CAZyme ID | MGYG000001537_01427 | |||||||||||
| CAZy Family | CE3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 331545; End: 332630 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE3 | 39 | 236 | 1.4e-63 | 0.9948453608247423 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd01833 | XynB_like | 1.78e-59 | 39 | 236 | 1 | 157 | SGNH_hydrolase subfamily, similar to Ruminococcus flavefaciens XynB. Most likely a secreted hydrolase with xylanase activity. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| pfam13472 | Lipase_GDSL_2 | 3.09e-19 | 43 | 221 | 1 | 171 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| cd00229 | SGNH_hydrolase | 9.72e-18 | 41 | 235 | 1 | 187 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
| pfam00657 | Lipase_GDSL | 7.38e-11 | 41 | 232 | 1 | 224 | GDSL-like Lipase/Acylhydrolase. |
| cd01831 | Endoglucanase_E_like | 1.42e-09 | 116 | 237 | 56 | 169 | Endoglucanase E-like members of the SGNH hydrolase family; Endoglucanase E catalyzes the endohydrolysis of 1,4-beta-glucosidic linkages in cellulose, lichenin and cereal beta-D-glucans. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCJ77184.1 | 7.11e-116 | 4 | 252 | 3 | 246 |
| AEE45464.1 | 1.64e-115 | 8 | 245 | 7 | 241 |
| VEH29483.1 | 1.64e-115 | 8 | 245 | 7 | 241 |
| BCB83596.1 | 7.78e-115 | 17 | 247 | 9 | 244 |
| BCB78724.1 | 5.06e-114 | 30 | 247 | 28 | 244 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5B5S_A | 7.50e-33 | 39 | 244 | 2 | 207 | Crystalstructure of a carbohydrate esterase family 3 from Talaromyces cellulolyticus [Talaromyces cellulolyticus] |
| 5B5L_A | 2.05e-32 | 39 | 244 | 2 | 207 | Crystalstructure of acetyl esterase mutant S10A with acetate ion [Talaromyces cellulolyticus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9RLB8 | 1.96e-14 | 21 | 253 | 25 | 278 | Multidomain esterase OS=Ruminococcus flavefaciens OX=1265 GN=cesA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
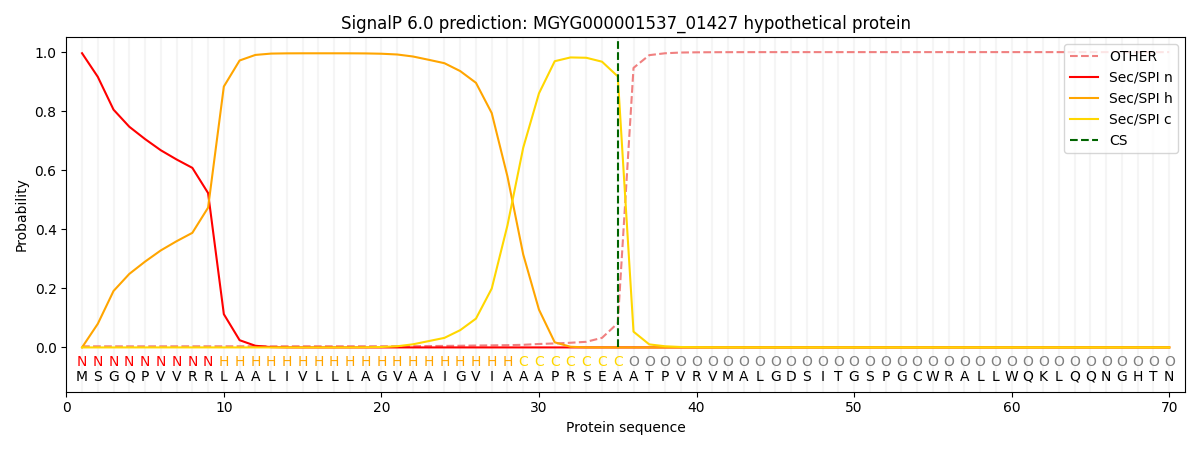
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.004905 | 0.994160 | 0.000244 | 0.000275 | 0.000181 | 0.000186 |