You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001537_01989
You are here: Home > Sequence: MGYG000001537_01989
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
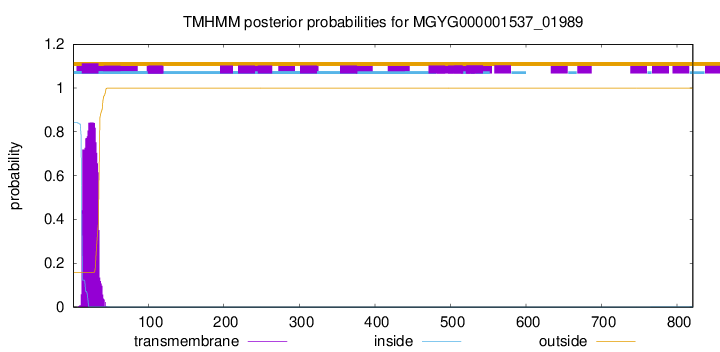
TMHMM annotations
Basic Information help
| Species | Cellulomonas timonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Cellulomonadaceae; Cellulomonas; Cellulomonas timonensis | |||||||||||
| CAZyme ID | MGYG000001537_01989 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 312595; End: 315060 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH62 | 504 | 788 | 1.1e-116 | 0.9928057553956835 |
| GH10 | 42 | 331 | 1.1e-108 | 0.9801980198019802 |
| CBM2 | 368 | 468 | 1.7e-36 | 0.9900990099009901 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd08987 | GH62 | 2.37e-173 | 505 | 816 | 1 | 304 | Glycosyl hydrolase family 62, characterized arabinofuranosidases. The glycosyl hydrolase family 62 (GH62) includes eukaryotic (mostly fungal) and prokaryotic enzymes which are characterized arabinofuranosidases (alpha-L-arabinofuranosidases; EC 3.2.1.55) that specifically cleave either alpha-1,2 or alpha-1,3-L-arabinofuranose side chains from xylans. These enzymes show significantly different substrate preference with rather low specific activity towards natural substrates and differ in catalytic efficiency. They do not act on xylose moieties in xylan that are adorned with an arabinose side chain at both O2 and O3 positions, nor do they display any non-specific arabinofuranosidase activity. The synergistic action in biomass degradation makes GH62 promising candidates for biotechnological improvements of biofuel production and in various biorefinery applications. These enzymes also contain carbohydrate binding modules (CBMs) that bind cellulose or xylan. |
| pfam00331 | Glyco_hydro_10 | 3.25e-138 | 39 | 331 | 1 | 310 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 1.42e-114 | 81 | 328 | 1 | 262 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 3.14e-77 | 38 | 326 | 23 | 334 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam03664 | Glyco_hydro_62 | 6.89e-60 | 505 | 788 | 3 | 272 | Glycosyl hydrolase family 62. Family of alpha -L-arabinofuranosidase (EC 3.2.1.55). This enzyme hydrolyzed aryl alpha-L-arabinofuranosides and cleaves arabinosyl side chains from arabinoxylan and arabinan. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QVI64496.1 | 0.0 | 1 | 821 | 1 | 814 |
| QCB92556.1 | 0.0 | 4 | 821 | 4 | 833 |
| QHT57555.1 | 1.20e-309 | 1 | 815 | 1 | 806 |
| ADG75907.1 | 7.40e-306 | 1 | 815 | 1 | 826 |
| QVI61811.1 | 7.92e-306 | 1 | 815 | 1 | 818 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5M0K_A | 9.35e-179 | 1 | 335 | 2 | 337 | CRYSTALSTRUCTURE of endo-1,4-beta-xylanase from Cellulomonas flavigena [Cellulomonas flavigena],5M0K_B CRYSTAL STRUCTURE of endo-1,4-beta-xylanase from Cellulomonas flavigena [Cellulomonas flavigena] |
| 5B6S_A | 1.48e-159 | 505 | 821 | 1 | 316 | ChainA, Glycosyl hydrolase family 62 protein [Coprinopsis cinerea okayama7#130],5B6S_B Chain B, Glycosyl hydrolase family 62 protein [Coprinopsis cinerea okayama7#130],5B6T_A Chain A, Glycosyl hydrolase family 62 protein [Coprinopsis cinerea okayama7#130],5B6T_B Chain B, Glycosyl hydrolase family 62 protein [Coprinopsis cinerea okayama7#130] |
| 1ISV_A | 8.00e-141 | 37 | 337 | 3 | 304 | Crystalstructure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylose [Streptomyces olivaceoviridis],1ISV_B Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylose [Streptomyces olivaceoviridis],1ISW_A Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylobiose [Streptomyces olivaceoviridis],1ISW_B Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylobiose [Streptomyces olivaceoviridis],1ISX_A Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylotriose [Streptomyces olivaceoviridis],1ISX_B Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylotriose [Streptomyces olivaceoviridis],1ISY_A Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with glucose [Streptomyces olivaceoviridis],1ISY_B Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with glucose [Streptomyces olivaceoviridis],1ISZ_A Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with galactose [Streptomyces olivaceoviridis],1ISZ_B Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with galactose [Streptomyces olivaceoviridis],1IT0_A Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with lactose [Streptomyces olivaceoviridis],1IT0_B Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with lactose [Streptomyces olivaceoviridis],1V6U_A Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 2(2)-alpha-L-arabinofuranosyl-xylobiose [Streptomyces olivaceoviridis],1V6U_B Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 2(2)-alpha-L-arabinofuranosyl-xylobiose [Streptomyces olivaceoviridis],1V6V_A Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 3(2)-alpha-L-arabinofuranosyl-xylotriose [Streptomyces olivaceoviridis],1V6V_B Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 3(2)-alpha-L-arabinofuranosyl-xylotriose [Streptomyces olivaceoviridis],1V6W_A Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 2(2)-4-O-methyl-alpha-D-glucuronosyl-xylobiose [Streptomyces olivaceoviridis],1V6W_B Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 2(2)-4-O-methyl-alpha-D-glucuronosyl-xylobiose [Streptomyces olivaceoviridis],1V6X_A Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 3(3)-4-O-methyl-alpha-D-glucuronosyl-xylotriose [Streptomyces olivaceoviridis],1V6X_B Crystal Structure Of Xylanase From Streptomyces Olivaceoviridis E-86 Complexed With 3(3)-4-O-methyl-alpha-D-glucuronosyl-xylotriose [Streptomyces olivaceoviridis],1XYF_A Endo-1,4-Beta-Xylanase From Streptomyces Olivaceoviridis [Streptomyces olivaceoviridis],1XYF_B Endo-1,4-Beta-Xylanase From Streptomyces Olivaceoviridis [Streptomyces olivaceoviridis] |
| 1E0W_A | 1.01e-140 | 37 | 334 | 3 | 301 | Xylanase10A from Sreptomyces lividans. native structure at 1.2 angstrom resolution [Streptomyces lividans],1E0X_A Xylanase 10a From Sreptomyces Lividans. Xylobiosyl-Enzyme Intermediate At 1.65 A [Streptomyces lividans],1E0X_B Xylanase 10a From Sreptomyces Lividans. Xylobiosyl-Enzyme Intermediate At 1.65 A [Streptomyces lividans],1OD8_A Xylanase Xyn10A from Streptomyces lividans in complex with xylobio-isofagomine lactam [Streptomyces lividans],1V0K_A Xylanase Xyn10A from Streptomyces lividans in complex with xylobio-deoxynojirimycin at pH 5.8 [Streptomyces lividans],1V0L_A Xylanase Xyn10A from Streptomyces lividans in complex with xylobio-isofagomine at pH 5.8 [Streptomyces lividans],1V0M_A Xylanase Xyn10a from Streptomyces lividans in complex with xylobio-deoxynojirimycin at pH 7.5 [Streptomyces lividans],1V0N_A Xylanase Xyn10a from Streptomyces lividans in complex with xylobio-isofagomine at pH 7.5 [Streptomyces lividans] |
| 2G3I_A | 1.01e-140 | 37 | 334 | 3 | 301 | Structureof S.olivaceoviridis xylanase Q88A/R275A mutant [Streptomyces olivaceoviridis],2G3J_A Structure of S.olivaceoviridis xylanase Q88A/R275A mutant [Streptomyces olivaceoviridis],2G4F_A Chain A, Hydrolase [Streptomyces olivaceoviridis],2G4F_B Chain B, Hydrolase [Streptomyces olivaceoviridis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P07986 | 2.29e-145 | 6 | 468 | 13 | 481 | Exoglucanase/xylanase OS=Cellulomonas fimi OX=1708 GN=cex PE=1 SV=1 |
| P26514 | 2.42e-139 | 32 | 334 | 39 | 342 | Endo-1,4-beta-xylanase A OS=Streptomyces lividans OX=1916 GN=xlnA PE=1 SV=2 |
| B4XVN1 | 4.38e-109 | 5 | 449 | 6 | 442 | Endo-1,4-beta-xylanase A OS=Streptomyces sp. OX=1931 GN=xynAS9 PE=1 SV=1 |
| P23031 | 3.64e-80 | 505 | 814 | 323 | 615 | Alpha-L-arabinofuranosidase C OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=xynC PE=1 SV=2 |
| Q0H904 | 1.35e-76 | 48 | 333 | 40 | 325 | Endo-1,4-beta-xylanase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=xlnC PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
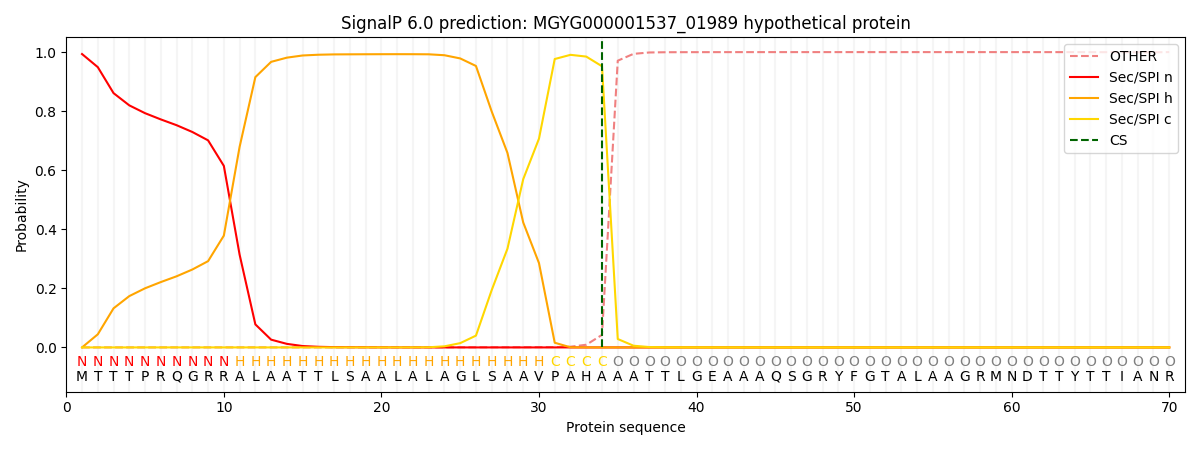
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000819 | 0.991098 | 0.000327 | 0.007136 | 0.000338 | 0.000258 |