You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000001537_02554
You are here: Home > Sequence: MGYG000001537_02554
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cellulomonas timonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Actinobacteriota; Actinomycetia; Actinomycetales; Cellulomonadaceae; Cellulomonas; Cellulomonas timonensis | |||||||||||
| CAZyme ID | MGYG000001537_02554 | |||||||||||
| CAZy Family | CBM2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 889153; End: 893292 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM2 | 1275 | 1376 | 6.8e-30 | 0.9900990099009901 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033681 | ExeM_NucH_DNase | 0.0 | 78 | 633 | 1 | 544 | ExeM/NucH family extracellular endonuclease. |
| COG2374 | COG2374 | 1.16e-109 | 5 | 645 | 146 | 798 | Predicted extracellular nuclease [General function prediction only]. |
| COG0737 | UshA | 1.80e-73 | 642 | 1160 | 24 | 503 | 2',3'-cyclic-nucleotide 2'-phosphodiesterase/5'- or 3'-nucleotidase, 5'-nucleotidase family [Nucleotide transport and metabolism, Defense mechanisms]. |
| PRK09419 | PRK09419 | 5.50e-71 | 637 | 1166 | 653 | 1143 | multifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase/5'-nucleotidase. |
| NF033680 | exonuc_ExeM-GG | 1.19e-62 | 59 | 632 | 200 | 843 | extracellular exonuclease ExeM. ExeM, as described in Shewanella oneidensis, is a biofilm formation-associated exonuclease that cleaves extracellular DNA (eDNA), a biofilm component. Members of the ExeM family contain two or three pairs of Cys residues, presumed to form disulfide bonds, and a C-terminal GlyGly-CTERM membrane-anchoring segment. Strangely, engineered removal of the GlyGly-CTERM region did not result in net export from the cell and appearance of the enzyme in culture supernatants. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QAY73698.1 | 1.34e-260 | 56 | 1270 | 297 | 1524 |
| QAY64865.1 | 7.24e-254 | 56 | 1271 | 245 | 1481 |
| SDS92078.1 | 4.91e-249 | 59 | 1339 | 241 | 1553 |
| ACZ32093.1 | 2.51e-247 | 56 | 1347 | 229 | 1564 |
| QAY71724.1 | 1.05e-244 | 56 | 1193 | 236 | 1404 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2Z1A_A | 1.47e-33 | 639 | 1152 | 24 | 514 | Crystalstructure of 5'-nucleotidase precursor from Thermus thermophilus HB8 [Thermus thermophilus HB8] |
| 3IVD_A | 1.39e-27 | 645 | 1158 | 7 | 487 | Putative5'-Nucleotidase (c4898) from Escherichia Coli in complex with Uridine [Escherichia coli O6],3IVD_B Putative 5'-Nucleotidase (c4898) from Escherichia Coli in complex with Uridine [Escherichia coli O6],3IVE_A Putative 5'-Nucleotidase (c4898) from Escherichia Coli in complex with Cytidine [Escherichia coli O6] |
| 5H7W_A | 2.28e-27 | 641 | 1167 | 1 | 523 | Crystalstructure of 5'-nucleotidase from venom of Naja atra [Naja atra],5H7W_B Crystal structure of 5'-nucleotidase from venom of Naja atra [Naja atra] |
| 7D0V_A | 4.10e-26 | 641 | 1167 | 1 | 523 | ChainA, Snake venom 5'-nucleotidase [Naja atra],7D0V_B Chain B, Snake venom 5'-nucleotidase [Naja atra] |
| 7QGM_A | 2.15e-23 | 628 | 1165 | 12 | 545 | ChainA, 5'-nucleotidase [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8YAJ5 | 4.88e-38 | 620 | 1171 | 7 | 581 | Cell wall protein Lmo0130 OS=Listeria monocytogenes serovar 1/2a (strain ATCC BAA-679 / EGD-e) OX=169963 GN=lmo0130 PE=1 SV=1 |
| P54602 | 1.90e-36 | 645 | 1152 | 590 | 1068 | Endonuclease YhcR OS=Bacillus subtilis (strain 168) OX=224308 GN=yhcR PE=1 SV=1 |
| A9BJC1 | 2.83e-29 | 651 | 1132 | 30 | 462 | Mannosylglucosyl-3-phosphoglycerate phosphatase OS=Petrotoga mobilis (strain DSM 10674 / SJ95) OX=403833 GN=mggB PE=1 SV=1 |
| A0A2I4HXH5 | 1.25e-26 | 641 | 1167 | 1 | 523 | Snake venom 5'-nucleotidase (Fragment) OS=Naja atra OX=8656 PE=1 SV=1 |
| B6EWW8 | 8.49e-26 | 621 | 1167 | 15 | 562 | Snake venom 5'-nucleotidase OS=Gloydius brevicaudus OX=259325 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
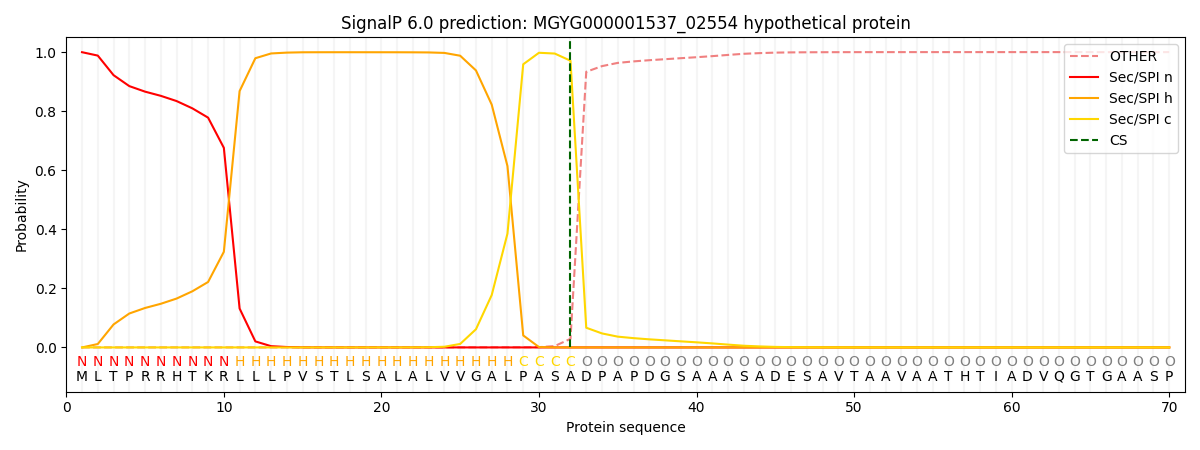
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000607 | 0.998264 | 0.000437 | 0.000293 | 0.000191 | 0.000163 |
